CopciAB_455101
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_455101 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 455101 |
| Uniprot id | Functional description | ||
| Location | scaffold_6:2212111..2214956 | Strand | - |
| Gene length (nt) | 2846 | Transcript length (nt) | 2614 |
| CDS length (nt) | 2289 | Protein length (aa) | 762 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7269054 | 31.6 | 1.848E-86 | 304 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB15192 | 30.7 | 1.777E-75 | 271 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1032201 | 30.7 | 2.763E-75 | 270 |
| Pleurotus ostreatus PC9 | PleosPC9_1_25429 | 30.5 | 1.195E-73 | 265 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1496586 | 32.1 | 5.349E-68 | 248 |
| Schizophyllum commune H4-8 | Schco3_2693547 | 27.4 | 2.502E-55 | 209 |
| Flammulina velutipes | Flave_chr06AA00915 | 26.2 | 1.898E-33 | 139 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15708 | 40 | 1.925E-14 | 77 |
| Lentinula edodes NBRC 111202 | Lenedo1_1140240 | 40 | 3.913E-14 | 76 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_455101.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 187 | 376 |
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 390 | 522 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
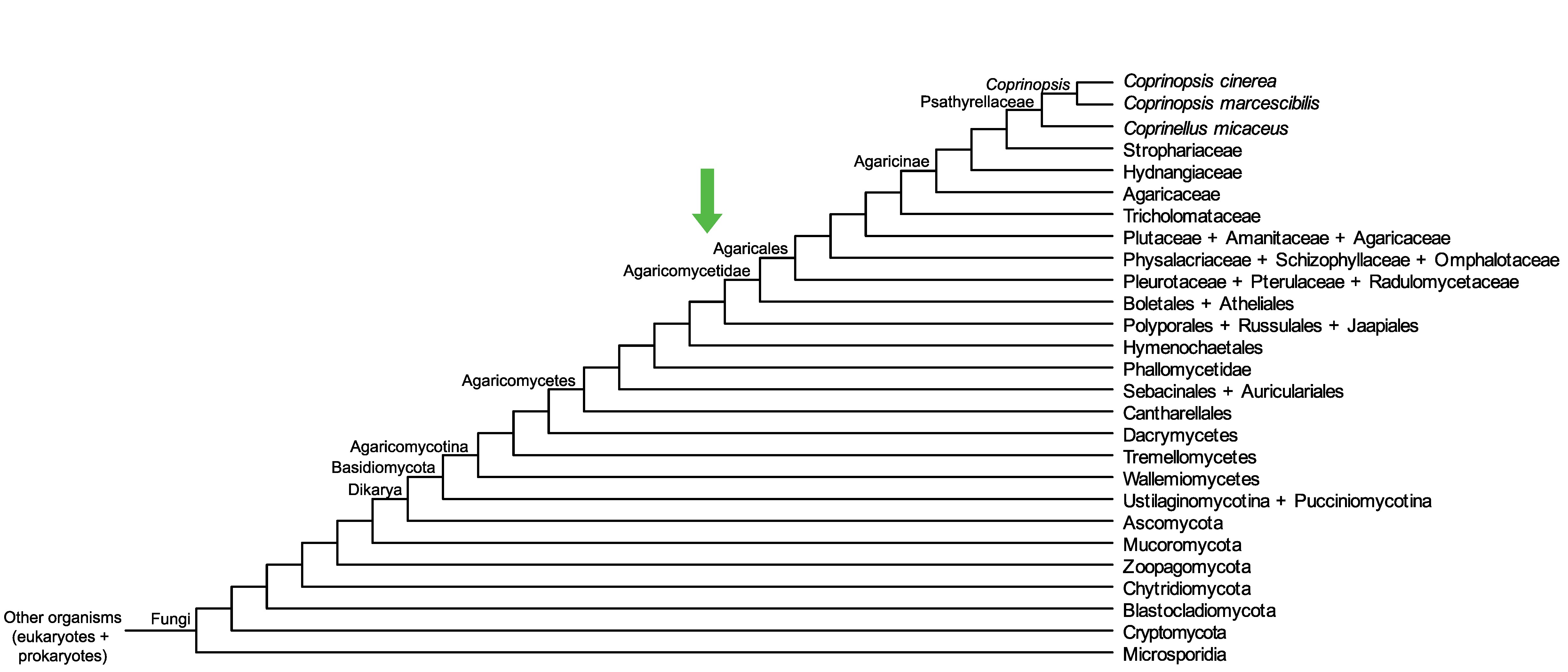
Conservation of CopciAB_455101 across fungi.
Arrow shows the origin of gene family containing CopciAB_455101.
Protein
| Sequence id | CopciAB_455101.T0 |
|---|---|
| Sequence |
>CopciAB_455101.T0 MAREVESNIIGPMPVETFLDEFVRSEKGLKLACSDQYPASFFDTVPTTATDTQRNSWKSARKTRQAKFKKCGKEK GMAEPWARRLNTLMPQYKSVNSSDSPDAQSDWDKKWRPDVSTYKKPEKGSYGASKILKGFKETQLDRVELVTEIK TTKNAVQAFQDAVHKNANPKEHEFLLDGKGAELLRGKVATMATEVLNRQHRTCFFMVLITDPHARLIRVDRAGIV VSAAIQFRIDPLPLIDFFLHFNNLSREGRGLDPTVRDPTLKEQAIAHEKLSAPKDGRGVKVIAVPGNGQGESSRE VLAYRPMVDAASLTGRCTRTYKVWDLKDNVFRFLKDTWRPDTEDSEKESAVLRELKEHGVPHIPDLICGDDIEGN VTRTQDFLDAPWVGARKRKPIDPRVNHRLLENLVPRFLEDCKNIRGVFQVIYDALVAHQKAYECGWLHRDISDNN VLVLEFEDRGVLADWELAIRVKDRNDEPVDRSKRPHYRTGTWDFMSIKLLSSLDAAHTVLDDLESFFWLVLHVSI RVVPIQSRLADVNTIMKDLFHASSWDSNTRSFFGGGYKEHCLLIDGGSTSFSNLTFPGNKALDSWYKKVRLALRA SFTRSASSEEQSGFNHDFLAQQFTTALESTEWSKEPLMGSESRVVVKAKGGPTTAARGAGKRDLATMQGSDDDVA QHGEEDDDENQGGDGDCDGDAEEGQGEADDDEGEDEEAEDEEAEGMDESEGEGPEDEDEGADGSGEPPMKSLRLD AQGTRRSTRRTA |
| Length | 762 |
Coding
| Sequence id | CopciAB_455101.T0 |
|---|---|
| Sequence |
>CopciAB_455101.T0 ATGGCAAGGGAAGTGGAATCCAACATCATAGGCCCTATGCCCGTCGAGACCTTTCTTGATGAATTCGTCCGAAGC GAGAAGGGATTGAAGCTGGCTTGCAGCGACCAATACCCTGCCTCGTTTTTTGACACTGTCCCCACCACCGCCACT GATACCCAACGAAACAGTTGGAAAAGTGCGAGAAAAACGCGACAGGCCAAGTTCAAGAAATGCGGAAAGGAAAAG GGAATGGCTGAACCCTGGGCCCGTAGACTTAATACCCTGATGCCCCAGTACAAATCGGTCAACTCGTCTGATTCG CCCGACGCCCAATCCGACTGGGATAAGAAGTGGCGACCGGACGTTTCCACCTACAAGAAGCCAGAAAAGGGCTCC TATGGGGCGAGTAAAATACTCAAGGGTTTTAAGGAGACCCAGCTGGATAGGGTCGAACTTGTCACAGAGATCAAA ACCACCAAGAATGCAGTTCAGGCCTTCCAAGACGCCGTTCACAAGAATGCCAACCCGAAGGAGCACGAGTTTCTC CTCGATGGTAAAGGGGCAGAGCTTTTGCGTGGAAAGGTGGCGACCATGGCTACGGAAGTTCTCAACCGCCAACAT CGCACTTGCTTCTTCATGGTCCTCATCACGGACCCGCACGCCCGCCTTATTCGTGTCGACAGGGCAGGTATCGTT GTCTCTGCAGCAATCCAATTCCGCATCGACCCCCTTCCACTCATCGACTTCTTTCTCCACTTCAACAACCTCTCA AGAGAGGGTCGCGGACTAGACCCGACCGTGCGTGATCCTACCCTCAAGGAACAGGCCATCGCCCACGAAAAGCTC TCAGCCCCGAAGGATGGACGCGGAGTGAAGGTCATCGCGGTTCCTGGTAACGGACAGGGCGAGTCTTCTCGCGAG GTGCTCGCCTATCGACCCATGGTGGACGCAGCATCTTTGACAGGGCGGTGCACCAGAACATACAAGGTCTGGGAC CTCAAAGACAATGTCTTTCGTTTTTTGAAGGATACGTGGCGGCCTGACACGGAGGATTCGGAGAAGGAGTCTGCC GTCTTAAGGGAGCTGAAGGAGCACGGCGTCCCTCACATCCCCGACCTGATATGTGGCGACGATATTGAAGGCAAC GTGACACGAACACAAGATTTCCTCGACGCTCCTTGGGTCGGCGCACGGAAACGCAAACCTATCGATCCACGGGTT AATCATCGGTTGCTCGAGAATCTAGTGCCACGTTTTCTCGAAGACTGCAAAAATATCCGTGGTGTCTTCCAAGTC ATATACGATGCGCTTGTCGCTCACCAGAAGGCCTATGAATGCGGGTGGTTGCACCGCGACATTAGCGACAATAAC GTTTTGGTTCTTGAATTCGAGGACAGAGGTGTCCTTGCCGACTGGGAGCTTGCCATTCGCGTCAAGGATAGAAAT GACGAACCCGTCGATCGGTCGAAACGTCCTCATTATAGAACCGGCACATGGGATTTTATGTCGATCAAGCTCCTC AGTTCATTGGACGCCGCTCACACTGTCCTTGACGACTTGGAGTCTTTCTTTTGGCTCGTCCTGCATGTCAGCATA CGAGTCGTCCCCATTCAGTCGCGCTTAGCGGACGTGAATACTATTATGAAAGATCTCTTTCACGCCTCCTCCTGG GATTCTAACACCCGTTCATTCTTTGGCGGCGGTTACAAAGAACACTGTCTGCTCATCGATGGAGGGTCCACCTCC TTCAGCAATCTGACATTCCCCGGCAACAAGGCACTTGATTCGTGGTATAAAAAGGTGCGACTGGCACTCCGCGCC TCCTTTACACGGTCAGCATCCTCAGAAGAACAATCTGGTTTTAATCACGACTTCCTGGCTCAACAATTCACAACT GCGCTAGAGAGTACAGAATGGTCGAAAGAACCACTCATGGGAAGTGAAAGCAGGGTCGTGGTAAAAGCGAAGGGT GGTCCAACGACAGCCGCCCGGGGAGCTGGGAAGCGCGATTTGGCGACTATGCAGGGCAGTGATGATGATGTTGCC CAGCATGGCGAGGAGGACGATGACGAGAACCAGGGTGGGGATGGGGACTGTGACGGGGATGCTGAGGAAGGTCAA GGGGAGGCAGACGACGACGAGGGAGAGGACGAAGAAGCAGAGGACGAAGAAGCAGAGGGCATGGATGAGAGCGAA GGCGAAGGGCCAGAAGATGAAGATGAGGGTGCGGATGGTAGTGGAGAGCCACCTATGAAGAGTCTGAGGTTAGAT GCCCAAGGTACCCGTCGCAGCACGCGCAGAACGGCCTGA |
| Length | 2289 |
Transcript
| Sequence id | CopciAB_455101.T0 |
|---|---|
| Sequence |
>CopciAB_455101.T0 AGCTTGAACACGAGCTGCCTCTGACCACAACCTGCCTCGAGCCACGTTTTGGGACTCATCTTGCCTCATTTAACT GGATCTCGTTTCACCATATAACCAGCGTACCCCCACCAAGGGCAAAAAGCTCTCTAGCCAGAGTCGGTACGAGCG CACCAGATCGCGCACACAAGAAGGAAGCGCCCTCGGCCAACAGGAGTTTCTCAAAGCAGTCGACGACTTACACGT CCAAATGGCAAGGGAAGTGGAATCCAACATCATAGGCCCTATGCCCGTCGAGACCTTTCTTGATGAATTCGTCCG AAGCGAGAAGGGATTGAAGCTGGCTTGCAGCGACCAATACCCTGCCTCGTTTTTTGACACTGTCCCCACCACCGC CACTGATACCCAACGAAACAGTTGGAAAAGTGCGAGAAAAACGCGACAGGCCAAGTTCAAGAAATGCGGAAAGGA AAAGGGAATGGCTGAACCCTGGGCCCGTAGACTTAATACCCTGATGCCCCAGTACAAATCGGTCAACTCGTCTGA TTCGCCCGACGCCCAATCCGACTGGGATAAGAAGTGGCGACCGGACGTTTCCACCTACAAGAAGCCAGAAAAGGG CTCCTATGGGGCGAGTAAAATACTCAAGGGTTTTAAGGAGACCCAGCTGGATAGGGTCGAACTTGTCACAGAGAT CAAAACCACCAAGAATGCAGTTCAGGCCTTCCAAGACGCCGTTCACAAGAATGCCAACCCGAAGGAGCACGAGTT TCTCCTCGATGGTAAAGGGGCAGAGCTTTTGCGTGGAAAGGTGGCGACCATGGCTACGGAAGTTCTCAACCGCCA ACATCGCACTTGCTTCTTCATGGTCCTCATCACGGACCCGCACGCCCGCCTTATTCGTGTCGACAGGGCAGGTAT CGTTGTCTCTGCAGCAATCCAATTCCGCATCGACCCCCTTCCACTCATCGACTTCTTTCTCCACTTCAACAACCT CTCAAGAGAGGGTCGCGGACTAGACCCGACCGTGCGTGATCCTACCCTCAAGGAACAGGCCATCGCCCACGAAAA GCTCTCAGCCCCGAAGGATGGACGCGGAGTGAAGGTCATCGCGGTTCCTGGTAACGGACAGGGCGAGTCTTCTCG CGAGGTGCTCGCCTATCGACCCATGGTGGACGCAGCATCTTTGACAGGGCGGTGCACCAGAACATACAAGGTCTG GGACCTCAAAGACAATGTCTTTCGTTTTTTGAAGGATACGTGGCGGCCTGACACGGAGGATTCGGAGAAGGAGTC TGCCGTCTTAAGGGAGCTGAAGGAGCACGGCGTCCCTCACATCCCCGACCTGATATGTGGCGACGATATTGAAGG CAACGTGACACGAACACAAGATTTCCTCGACGCTCCTTGGGTCGGCGCACGGAAACGCAAACCTATCGATCCACG GGTTAATCATCGGTTGCTCGAGAATCTAGTGCCACGTTTTCTCGAAGACTGCAAAAATATCCGTGGTGTCTTCCA AGTCATATACGATGCGCTTGTCGCTCACCAGAAGGCCTATGAATGCGGGTGGTTGCACCGCGACATTAGCGACAA TAACGTTTTGGTTCTTGAATTCGAGGACAGAGGTGTCCTTGCCGACTGGGAGCTTGCCATTCGCGTCAAGGATAG AAATGACGAACCCGTCGATCGGTCGAAACGTCCTCATTATAGAACCGGCACATGGGATTTTATGTCGATCAAGCT CCTCAGTTCATTGGACGCCGCTCACACTGTCCTTGACGACTTGGAGTCTTTCTTTTGGCTCGTCCTGCATGTCAG CATACGAGTCGTCCCCATTCAGTCGCGCTTAGCGGACGTGAATACTATTATGAAAGATCTCTTTCACGCCTCCTC CTGGGATTCTAACACCCGTTCATTCTTTGGCGGCGGTTACAAAGAACACTGTCTGCTCATCGATGGAGGGTCCAC CTCCTTCAGCAATCTGACATTCCCCGGCAACAAGGCACTTGATTCGTGGTATAAAAAGGTGCGACTGGCACTCCG CGCCTCCTTTACACGGTCAGCATCCTCAGAAGAACAATCTGGTTTTAATCACGACTTCCTGGCTCAACAATTCAC AACTGCGCTAGAGAGTACAGAATGGTCGAAAGAACCACTCATGGGAAGTGAAAGCAGGGTCGTGGTAAAAGCGAA GGGTGGTCCAACGACAGCCGCCCGGGGAGCTGGGAAGCGCGATTTGGCGACTATGCAGGGCAGTGATGATGATGT TGCCCAGCATGGCGAGGAGGACGATGACGAGAACCAGGGTGGGGATGGGGACTGTGACGGGGATGCTGAGGAAGG TCAAGGGGAGGCAGACGACGACGAGGGAGAGGACGAAGAAGCAGAGGACGAAGAAGCAGAGGGCATGGATGAGAG CGAAGGCGAAGGGCCAGAAGATGAAGATGAGGGTGCGGATGGTAGTGGAGAGCCACCTATGAAGAGTCTGAGGTT AGATGCCCAAGGTACCCGTCGCAGCACGCGCAGAACGGCCTGATGCCTCCTTACATTCCCCTATATACCATGTAG TATTATTCCTATATCCTAATACCTTAGTCAGTTCGTCGCGAGGGCCGCCGATCTTACTCCATTC |
| Length | 2614 |
Gene
| Sequence id | CopciAB_455101.T0 |
|---|---|
| Sequence |
>CopciAB_455101.T0 AGCTTGAACACGAGCTGCCTCTGACCACAACCTGCCTCGAGCCACGTTTTGGGACTCATCTTGCCTCATTTAACT GGATCTCGTTTCACCATATAACCAGGTAAGCCACCTTCCAGGTCCTCAGCACTACCTCTCGTGATGTCCGATTCA AACATTCAACAGCGTACCCCCACCAAGGGCAAAAAGCTCTCTAGCCAGAGTCGGTACGAGCGCACCAGATCGCGC ACACAAGAAGGAAGCGCCCTCGGCCAACAGGAGTTTCTCAAAGCAGTCGACGACTTACACGTCCAAATGGCAAGG GAAGTGGAATCCAACATCATAGGCCCTATGCCCGTCGAGACCTTTCTTGATGAATTCGTCCGAAGCGAGAAGGGA TTGAAGCTGGCTTGCAGCGACCAATACCCTGCCTCGTTTTTTGACACTGTCCCCACCACCGCCACTGATACCCAA CGAAACAGTTGGAAAAGTGCGAGAAAAACGCGACAGGCCAAGTTCAAGAAATGCGGAAAGGAAAAGGGAATGGCT GAACCCTGGGTATGTCAAATCTCTGTGTCTGTTGATAGTCCGACTAAGCACCGCACCTTTGACCAGGCCCGTAGA CTTAATACCCTGATGCCCCAGTACAAATCGGTCAACTCGTCTGATTCGCCCGACGCCCAATCCGACTGGGATAAG AAGTGGCGACCGGACGTTTCCACCTACAAGAAGCCAGAAAAGGGCTCCTATGGGGCGAGTAAAATACTCAAGGGT TTTAAGGAGACCCAGCTGGATAGGGTCGAACTTGTCACAGAGATCAAAACCACCAAGAATGCAGTTCAGGCCTTC CAAGACGCCGTTCACAAGAATGCCAACCCGAAGGAGCACGAGTTTCTCCTCGATGGTAAAGGGGCAGAGCTTTTG CGTGGAAAGGTGGCGACCATGGCTACGGAAGTTCTCAACCGCCAACATCGCACTTGCTTCTTCATGGTCCTCATC ACGGACCCGCACGCCCGCCTTATTCGTGTCGACAGGGCAGGTATCGTTGTCTCTGCAGCAATCCAATTCCGCATC GACCCCCTTCCACTCATCGACTTCTTTCTCCACTTCAACAACCTCTCAAGAGAGGGTCGCGGACTAGACCCGACC GTGCGTGATCCTACCCTCAAGGAACAGGCCATCGCCCACGAAAAGCTCTCAGCCCCGAAGGATGGACGCGGAGTG AAGGTCATCGCGGTTCCTGGTAACGGACAGGGCGAGTCTTCTCGCGAGGTGCTCGCCTATCGACCCATGGTGGAC GCAGCATCTTTGACAGGGCGGTGCACCAGAACATACAAGGTCTGGGACCTCAAAGACAATGTCTTTCGTTTTTTG AAGGATACGTGGCGGCCTGACACGGAGGATTCGGAGAAGGAGTCTGCCGTCTTAAGGGAGCTGAAGGAGCACGGC GTCCCTCACATCCCCGACCTGATATGTGGCGACGATATTGAAGGCAACGTGACACGAACACAAGATTTCCTCGAC GCTCCTTGGGTCGGCGCACGGAAACGCAAACCTATCGATCCACGGGTTAATCATCGGTTGCTCGAGAATCTAGTG CCACGTTTTCTCGAAGACTGCAAAAATATCCGTGGTGTCTTCCAAGTCATATACGATGCGCTTGTCGGTGCGTAG CTCCAGCCGTGGCCTGAGTGGCTTCCATTGCCACTAATGCTTCTATATGCAGCTCACCAGAAGGCCTATGAATGC GGGTGGTTGCACCGCGACATTAGCGACAATAACGTTTTGGTTCTTGAATTCGAGGACAGAGGTGTCCTTGCCGAC TGGGAGCTTGCCATTCGCGTCAAGGATAGAAATGACGAACCCGTCGATCGGTCGAAACGTCCTCATTATAGAACC GTGAGTGAAACCGATCTTCTCACCGAAATTTGCTCCTGGACAGTCTTATCCAGGGCACATGGGATTTTATGTCGA TCAAGCTCCTCAGTTCATTGGACGCCGCTCACACTGTCCTTGACGACTTGGAGTCTTTCTTTTGGCTCGTCCTGC ATGTCAGCATACGAGTCGTCCCCATTCAGTCGCGCTTAGCGGACGTGAATACTATTATGAAAGATCTCTTTCACG CCTCCTCCTGGGATTCTAACACCCGTTCATTCTTTGGCGGCGGTTACAAAGAACACTGTCTGCTCATCGATGGAG GGTCCACCTCCTTCAGCAATCTGACATTCCCCGGCAACAAGGCACTTGATTCGTGGTATAAAAAGGTGCGACTGG CACTCCGCGCCTCCTTTACACGGTCAGCATCCTCAGAAGAACAATCTGGTTTTAATCACGACTTCCTGGCTCAAC AATTCACAACTGCGCTAGAGAGTACAGAATGGTCGAAAGAACCACTCATGGGAAGTGAAAGCAGGGTCGTGGTAA AAGCGAAGGGTGGTCCAACGACAGCCGCCCGGGGAGCTGGGAAGCGCGATTTGGCGACTATGCAGGGCAGTGATG ATGATGTTGCCCAGCATGGCGAGGAGGACGATGACGAGAACCAGGGTGGGGATGGGGACTGTGACGGGGATGCTG AGGAAGGTCAAGGGGAGGCAGACGACGACGAGGGAGAGGACGAAGAAGCAGAGGACGAAGAAGCAGAGGGCATGG ATGAGAGCGAAGGCGAAGGGCCAGAAGATGAAGATGAGGGTGCGGATGGTAGTGGAGAGCCACCTATGAAGAGTC TGAGGTTAGATGCCCAAGGTACCCGTCGCAGCACGCGCAGAACGGCCTGATGCCTCCTTACATTCCCCTATATAC CATGTAGTATTATTCCTATATCCTAATACCTTAGTCAGTTCGTCGCGAGGGCCGCCGATCTTACTCCATTC |
| Length | 2846 |