CopciAB_456025
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_456025 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 456025 |
| Uniprot id | Functional description | other FunK1 protein kinase | |
| Location | scaffold_6:2675010..2677967 | Strand | + |
| Gene length (nt) | 2958 | Transcript length (nt) | 2745 |
| CDS length (nt) | 2310 | Protein length (aa) | 769 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262391 | 31.8 | 3.738E-74 | 267 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_456025.T0 |
| Description | other FunK1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 231 | 371 |
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 476 | 604 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR040976 | Fungal-type protein kinase |
| IPR008266 | Tyrosine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | other FunK1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
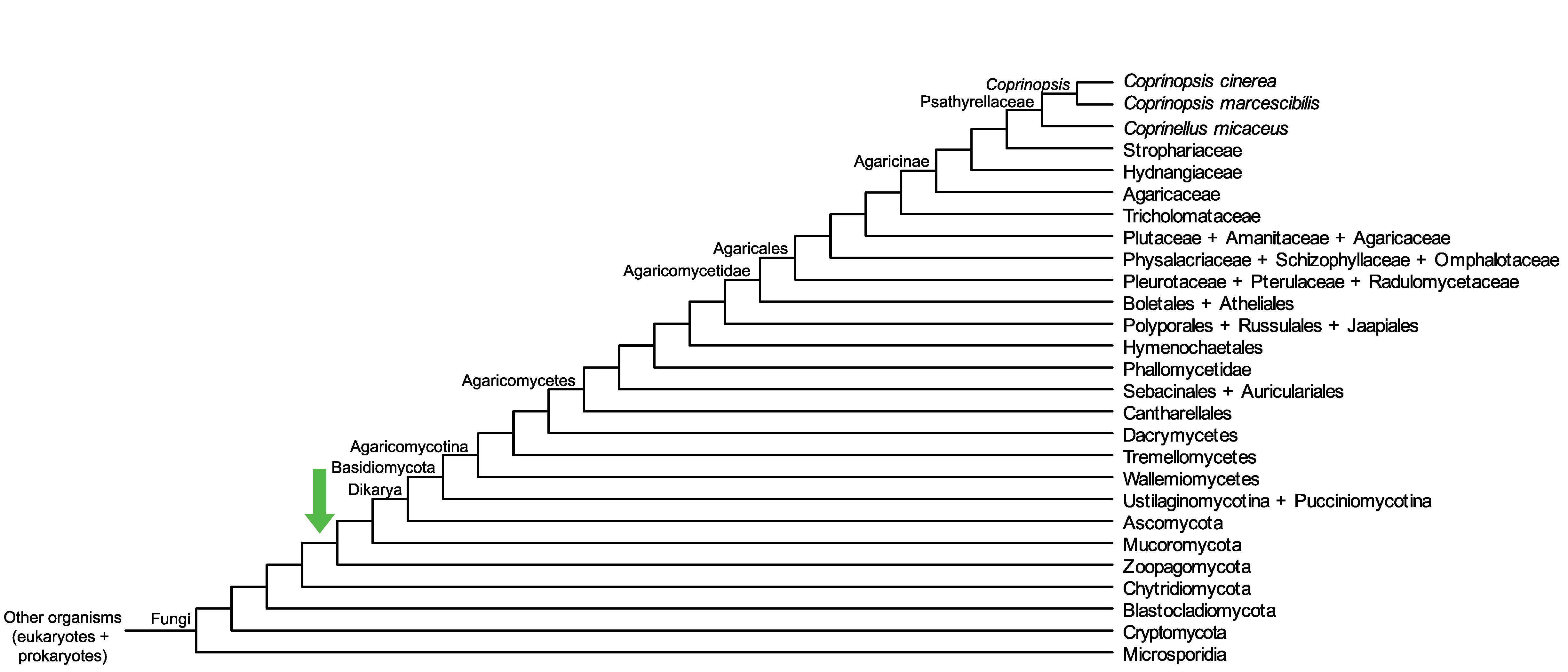
Conservation of CopciAB_456025 across fungi.
Arrow shows the origin of gene family containing CopciAB_456025.
Protein
| Sequence id | CopciAB_456025.T0 |
|---|---|
| Sequence |
>CopciAB_456025.T0 MTVAEQKAIIKAELAIATVASTDEDFPRQLFPDPGDDEIDAYLKATPLYDTESGRWVKLPKDVNCEEKLINIPMS EILGEILESFDLSAPNPGNRVVWDLTTKKSQRLAHEKEMAAKEKEEKDRVAREKQARPKSTKEKVELATDEGGDN GLDGKAEAATGKGQEDGAKETKRAKTFPKAMHHSEAVPTKAYSIPDTVFTGTGPGFTIDDVDMERPLYRASLGVV DYKRDSELDGSITENTAQLGVYARQVMNHQPNRNFVRAANVSNERVRLFHFDRSGVKYSLLYNIHSNPRMFVRII LGLVSTNLADLGFDPSIVWEPTDDPAAKFKRGTLTTRTANNDAIVYDIVGVEPFYRRRDLVGRGTLCWSIEIRTA SNRDQTEARNPDGQVEALFEKEGSAERDADVVEGEGRKSESEAADNGLSSKESWCSESRTSEEVFLAAAVGVKGV GQMVACEDVVETASFRGANAINDPGFLNRIKRRIILKKYGPPIHKFKTVLDVLYALIDAIRAHQRMVKEKGVLHR DISVNNILLGIAGADSEEGWRGVLIDFDMAIFLDRAEALLNELRTGTRMFQSLNVLQAWRPHDYLDDLESFLYVL IYLVLAFPEPNVECELPKELQDWNADSLYEAYKSKLTFITHGVLNFDVPPMWGYHISTLVEDLCRHFQPIHAMSI RISNQRLPKAQRDKLEASFKKKYLNDVDGQYDEIVGYIESAIENILAGQDGANEPSTMSSPSPTPAPRKGAGKRG SEEAYPHDAKEPSMKRSRH |
| Length | 769 |
Coding
| Sequence id | CopciAB_456025.T0 |
|---|---|
| Sequence |
>CopciAB_456025.T0 ATGACAGTCGCCGAGCAGAAGGCCATCATCAAAGCCGAGCTCGCGATCGCCACCGTCGCCTCCACTGACGAGGAT TTCCCTAGACAGCTCTTCCCTGACCCAGGCGACGATGAGATTGACGCCTATCTTAAGGCAACTCCACTGTACGAC ACGGAGAGTGGTCGATGGGTGAAGCTTCCGAAGGATGTCAACTGCGAGGAGAAGTTGATCAACATCCCAATGTCG GAGATTCTGGGAGAAATACTCGAGTCTTTCGATCTTTCTGCCCCCAATCCCGGCAATAGGGTGGTTTGGGATCTT ACGACTAAGAAATCACAGAGACTCGCGCACGAGAAGGAGATGGCGGCGAAAGAGAAGGAAGAGAAGGATAGGGTG GCACGGGAGAAGCAAGCGAGGCCGAAATCGACGAAGGAGAAGGTGGAACTGGCCACGGATGAAGGGGGAGACAAT GGGCTAGATGGTAAAGCGGAAGCAGCCACCGGGAAGGGCCAGGAGGATGGTGCGAAGGAAACGAAAAGAGCCAAA ACTTTCCCGAAAGCGATGCACCATTCGGAGGCCGTTCCAACGAAGGCGTATTCCATTCCTGATACGGTCTTCACT GGTACTGGTCCAGGCTTTACCATCGACGATGTTGACATGGAACGCCCTCTCTACCGGGCTTCCTTGGGTGTCGTG GACTACAAGCGGGATTCCGAACTCGATGGGTCGATCACTGAGAACACAGCTCAACTGGGTGTATACGCACGGCAA GTTATGAATCACCAACCCAACCGTAACTTCGTGCGTGCAGCGAACGTGAGCAACGAACGCGTTCGCCTTTTCCAT TTTGATCGCAGCGGCGTCAAGTACTCCTTGCTTTACAACATCCACAGCAACCCTCGCATGTTTGTACGCATTATC CTCGGCCTTGTCTCGACGAACCTAGCCGACCTTGGTTTCGATCCGTCGATCGTTTGGGAACCCACCGACGACCCT GCCGCAAAGTTCAAGCGCGGTACACTCACCACTCGAACCGCCAACAACGACGCAATTGTCTACGATATCGTCGGT GTGGAGCCTTTTTATCGACGACGCGACTTGGTTGGACGAGGAACACTTTGCTGGAGTATCGAGATCCGCACGGCA AGCAATCGCGACCAGACTGAGGCGAGAAATCCAGATGGGCAGGTTGAGGCGCTTTTCGAGAAGGAAGGATCGGCG GAGCGGGATGCAGACGTTGTCGAGGGTGAGGGGAGGAAGTCGGAATCGGAGGCGGCGGACAATGGGCTGTCCAGC AAGGAATCATGGTGCTCGGAATCGCGGACGTCGGAGGAGGTCTTTCTCGCAGCGGCTGTGGGTGTCAAGGGTGTC GGGCAGATGGTCGCATGTGAGGATGTGGTGGAAACTGCGAGCTTCAGGGGTGCCAATGCGATAAACGATCCTGGC TTCTTGAACCGAATCAAGCGACGCATCATCCTTAAGAAGTACGGACCGCCAATCCACAAGTTCAAGACGGTACTC GACGTACTCTATGCATTGATCGACGCCATTCGAGCCCACCAGCGTATGGTGAAGGAGAAAGGAGTTCTACACCGC GATATCAGTGTTAACAATATTCTATTGGGAATAGCTGGTGCCGACTCTGAGGAGGGATGGCGAGGAGTCCTTATT GACTTCGATATGGCGATCTTCCTTGATCGTGCTGAAGCGCTTTTGAATGAGCTCAGGACTGGTACGCGCATGTTC CAATCCCTCAATGTTCTCCAAGCATGGCGTCCACACGACTACTTGGACGACCTCGAGTCGTTCTTGTATGTTCTG ATTTACCTTGTCCTCGCCTTTCCTGAGCCCAACGTGGAATGCGAGCTGCCGAAGGAGCTCCAGGATTGGAACGCG GACAGTCTGTACGAAGCCTACAAGAGCAAGTTGACATTCATCACCCACGGCGTTTTGAACTTCGATGTGCCTCCG ATGTGGGGTTATCACATCTCTACGCTCGTCGAGGACCTATGTCGCCATTTCCAACCCATCCATGCCATGTCTATT CGCATTTCTAATCAAAGGCTACCCAAAGCCCAGCGCGACAAACTGGAAGCCTCGTTTAAGAAGAAGTATTTGAAC GACGTAGACGGGCAGTATGACGAGATTGTTGGATACATCGAGTCGGCCATCGAGAATATTCTTGCAGGCCAAGAC GGCGCTAATGAGCCTTCCACTATGTCCTCCCCCTCCCCCACTCCAGCCCCTCGAAAGGGCGCCGGGAAGAGGGGA TCTGAGGAAGCCTACCCCCATGACGCCAAGGAGCCCTCCATGAAGCGATCTCGTCACTAA |
| Length | 2310 |
Transcript
| Sequence id | CopciAB_456025.T0 |
|---|---|
| Sequence |
>CopciAB_456025.T0 CCTGACCACAACCATTGTGTCCTCGATTTGGACTGCTGGACGGCCAGCACCTATAAAGCATCACTCAAAGGGCAA GCCGTCTCCGACGCTACTCAACAACCAGCTGCGAGCACCACCTCGGAGCTCAGCGATACCGTTGGACCACTTGTC ACACTCCCGCCTCGAACGCCCAATCGAACGGTTCGACTTGCTTCCGGCTTTCAGTCTACCCCTCTGGCTGAGCGC AGTACCCAGTCCGTAGCCTACTCGACCACAATGACAGTCGCCGAGCAGAAGGCCATCATCAAAGCCGAGCTCGCG ATCGCCACCGTCGCCTCCACTGACGAGGATTTCCCTAGACAGCTCTTCCCTGACCCAGGCGACGATGAGATTGAC GCCTATCTTAAGGCAACTCCACTGTACGACACGGAGAGTGGTCGATGGGTGAAGCTTCCGAAGGATGTCAACTGC GAGGAGAAGTTGATCAACATCCCAATGTCGGAGATTCTGGGAGAAATACTCGAGTCTTTCGATCTTTCTGCCCCC AATCCCGGCAATAGGGTGGTTTGGGATCTTACGACTAAGAAATCACAGAGACTCGCGCACGAGAAGGAGATGGCG GCGAAAGAGAAGGAAGAGAAGGATAGGGTGGCACGGGAGAAGCAAGCGAGGCCGAAATCGACGAAGGAGAAGGTG GAACTGGCCACGGATGAAGGGGGAGACAATGGGCTAGATGGTAAAGCGGAAGCAGCCACCGGGAAGGGCCAGGAG GATGGTGCGAAGGAAACGAAAAGAGCCAAAACTTTCCCGAAAGCGATGCACCATTCGGAGGCCGTTCCAACGAAG GCGTATTCCATTCCTGATACGGTCTTCACTGGTACTGGTCCAGGCTTTACCATCGACGATGTTGACATGGAACGC CCTCTCTACCGGGCTTCCTTGGGTGTCGTGGACTACAAGCGGGATTCCGAACTCGATGGGTCGATCACTGAGAAC ACAGCTCAACTGGGTGTATACGCACGGCAAGTTATGAATCACCAACCCAACCGTAACTTCGTGCGTGCAGCGAAC GTGAGCAACGAACGCGTTCGCCTTTTCCATTTTGATCGCAGCGGCGTCAAGTACTCCTTGCTTTACAACATCCAC AGCAACCCTCGCATGTTTGTACGCATTATCCTCGGCCTTGTCTCGACGAACCTAGCCGACCTTGGTTTCGATCCG TCGATCGTTTGGGAACCCACCGACGACCCTGCCGCAAAGTTCAAGCGCGGTACACTCACCACTCGAACCGCCAAC AACGACGCAATTGTCTACGATATCGTCGGTGTGGAGCCTTTTTATCGACGACGCGACTTGGTTGGACGAGGAACA CTTTGCTGGAGTATCGAGATCCGCACGGCAAGCAATCGCGACCAGACTGAGGCGAGAAATCCAGATGGGCAGGTT GAGGCGCTTTTCGAGAAGGAAGGATCGGCGGAGCGGGATGCAGACGTTGTCGAGGGTGAGGGGAGGAAGTCGGAA TCGGAGGCGGCGGACAATGGGCTGTCCAGCAAGGAATCATGGTGCTCGGAATCGCGGACGTCGGAGGAGGTCTTT CTCGCAGCGGCTGTGGGTGTCAAGGGTGTCGGGCAGATGGTCGCATGTGAGGATGTGGTGGAAACTGCGAGCTTC AGGGGTGCCAATGCGATAAACGATCCTGGCTTCTTGAACCGAATCAAGCGACGCATCATCCTTAAGAAGTACGGA CCGCCAATCCACAAGTTCAAGACGGTACTCGACGTACTCTATGCATTGATCGACGCCATTCGAGCCCACCAGCGT ATGGTGAAGGAGAAAGGAGTTCTACACCGCGATATCAGTGTTAACAATATTCTATTGGGAATAGCTGGTGCCGAC TCTGAGGAGGGATGGCGAGGAGTCCTTATTGACTTCGATATGGCGATCTTCCTTGATCGTGCTGAAGCGCTTTTG AATGAGCTCAGGACTGGTACGCGCATGTTCCAATCCCTCAATGTTCTCCAAGCATGGCGTCCACACGACTACTTG GACGACCTCGAGTCGTTCTTGTATGTTCTGATTTACCTTGTCCTCGCCTTTCCTGAGCCCAACGTGGAATGCGAG CTGCCGAAGGAGCTCCAGGATTGGAACGCGGACAGTCTGTACGAAGCCTACAAGAGCAAGTTGACATTCATCACC CACGGCGTTTTGAACTTCGATGTGCCTCCGATGTGGGGTTATCACATCTCTACGCTCGTCGAGGACCTATGTCGC CATTTCCAACCCATCCATGCCATGTCTATTCGCATTTCTAATCAAAGGCTACCCAAAGCCCAGCGCGACAAACTG GAAGCCTCGTTTAAGAAGAAGTATTTGAACGACGTAGACGGGCAGTATGACGAGATTGTTGGATACATCGAGTCG GCCATCGAGAATATTCTTGCAGGCCAAGACGGCGCTAATGAGCCTTCCACTATGTCCTCCCCCTCCCCCACTCCA GCCCCTCGAAAGGGCGCCGGGAAGAGGGGATCTGAGGAAGCCTACCCCCATGACGCCAAGGAGCCCTCCATGAAG CGATCTCGTCACTAAGTTCCCGTCTTCGTATGTGATTCATCTTTTGGATCATCCCCTCTTTTACTGGTATCCTTA CACTTATCTATCCCCTGCCGTACTAATTCTATCTCATCCGTATATTCCTGACATCTAACATGTATCATCTTTGTA CTGTATACTTATACCAAATGGAAGTTTGGTTGTATTTTTAACCCC |
| Length | 2745 |
Gene
| Sequence id | CopciAB_456025.T0 |
|---|---|
| Sequence |
>CopciAB_456025.T0 CCTGACCACAACCATTGTGTCCTCGATTTGGACTGCTGGACGGCCAGCACCTATAAAGCATCACTCAAAGGGCAA GGTAAGTCCCCAGATTTGACTTGAAACTCGACTATGACCGCTAACAGACGGACCCCAGCCGTCTCCGACGCTACT CAACAACCAGCTGCGAGCACCACCTCGGAGCTCAGCGATACCGTTGGACCACTTGTCACACTCCCGCCTCGAACG CCCAATCGAACGGTTCGACTTGCTTCCGGCTTTCAGTCTACCCCTCTGGCTGAGCGCAGTACCCAGTCCGTAGCC TACTCGACCACAATGACAGTCGCCGAGCAGAAGGCCATCATCAAAGCCGAGCTCGCGATCGCCACCGTCGCCTCC ACTGACGAGGATTTCCCTAGACAGCTCTTCCCTGACCCAGGCGACGATGAGATTGACGCCTATCTTAAGGCAACT CCACTGTACGACACGGAGAGTGGTCGATGGGTGAAGCTTCCGAAGGATGTCAACTGCGAGGAGAAGTTGATCAAC ATCCCAATGTCGGAGATTCTGGGAGAAATACTCGAGTCTTTCGATCTTTCTGCCCCCAATCCCGGCAATAGGGTG GTTTGGGATCTTACGACTAAGAAATCACAGAGACTCGCGCACGAGAAGGAGATGGCGGCGAAAGAGAAGGAAGAG AAGGATAGGGTGGCACGGGAGAAGCAAGCGAGGCCGAAATCGACGAAGGAGAAGGTGGAACTGGCCACGGATGAA GGGGGAGACAATGGGCTAGATGGTAAAGCGGAAGCAGCCACCGGGAAGGGCCAGGAGGATGGTGCGAAGGAAACG AAAAGAGCCAAAACTTTCCCGAAAGCGATGCACCATTCGGAGGCCGTTCCAACGAAGGCGTATTCCATTCCTGAT ACGGTCTTCACTGGTACTGGTCCAGGCTTTACCATCGACGATGTTGACATGGAACGCCCTCTCTACCGGGCTTCC TTGGGTGTCGTGGACTACAAGCGGGATTCCGAACTCGATGGGTCGATCACTGAGAACACAGCTCAACTGGGTGTA TACGCACGGTAAGTCGTCCTATTCCTTCATCCGTGCTCCAGTCTGACCCCCTGCTAGGCAAGTTATGAATCACCA ACCCAACCGTAACTTCGTGCGTGCAGCGAACGTGAGCAACGAACGCGTTCGCCTTTTCCATTTTGATCGCAGCGG CGTCAAGTACTCCTTGCTTTACAACATCCACAGCAACCCTCGCATGTTTGTACGCATTATCCTCGGCCTTGTCTC GACGAACCTAGCCGACCTTGGTTTCGATCCGTCGATCGTTTGGGAACCCACCGACGACCCTGCCGCAAAGTTCAA GCGCGGTACACTCACCACTCGAACCGCCAACAACGACGCAATTGTCTACGATATCGTCGGTGTGGAGCCTTTTTA TCGACGACGCGACTTGGTTGGACGAGGAACACTTTGCTGGAGTATCGAGATCCGCACGGCAAGCAATCGCGACCA GACTGAGGCGAGAAATCCAGATGGGCAGGTTGAGGCGCTTTTCGAGAAGGAAGGATCGGCGGAGCGGGATGCAGA CGTTGTCGAGGGTGAGGGGAGGAAGTCGGAATCGGAGGCGGCGGACAATGGGCTGTCCAGCAAGGAATCATGGTG CTCGGAATCGCGGACGTCGGAGGAGGTCTTTCTCGCAGCGGCTGTGGGTGTCAAGGGTGTCGGGCAGATGGTCGC ATGTGAGGATGTGGTGGAAACTGCGAGCTTCAGGGGTGCCAATGCGATAAACGATCCTGGCTTCTTGAACCGAAT CAAGCGACGCATCATCCTTAAGAAGTACGGACCGCCAATCCACAAGTTCAAGACGGTACTCGACGTACTCTATGC ATTGATCGACGCCATTCGAGGTAAGTAAATTTTAACCGCCTCTGTTGGCACTCCCTGCTCACATTTTTTGTCTCA GCCCACCAGCGTATGGTGAAGGAGAAAGGAGTTCTACACCGCGATATCAGTGTTAACAATATTCTATTGGGAATA GCTGGTGCCGACTCTGAGGAGGGATGGCGAGGAGTCCTTATTGACTTCGATATGGCGATCTTCCTTGATCGTGCT GAAGCGCTTTTGAATGAGCTCAGGACTGTAAGTTTCGGCATTCTCTGCGGCCATATCTCTTACTGACCCCCGAGA AAGGGTACGCGCATGTTCCAATCCCTCAATGTTCTCCAAGCATGGCGTCCACACGACTACTTGGACGACCTCGAG TCGTTCTTGTATGTTCTGATTTACCTTGTCCTCGCCTTTCCTGAGCCCAACGTGGAATGCGAGCTGCCGAAGGAG CTCCAGGATTGGAACGCGGACAGTCTGTACGAAGCCTACAAGAGCAAGTTGACATTCATCACCCACGGCGTTTTG AACTTCGATGTGCCTCCGATGTGGGGTTATCACATCTCTACGCTCGTCGAGGACCTATGTCGCCATTTCCAACCC ATCCATGCCATGTCTATTCGCATTTCTAATCAAAGGCTACCCAAAGCCCAGCGCGACAAACTGGAAGCCTCGTTT AAGAAGAAGTATTTGAACGACGTAGACGGGCAGTATGACGAGATTGTTGGATACATCGAGTCGGCCATCGAGAAT ATTCTTGCAGGCCAAGACGGCGCTAATGAGCCTTCCACTATGTCCTCCCCCTCCCCCACTCCAGCCCCTCGAAAG GGCGCCGGGAAGAGGGGATCTGAGGAAGCCTACCCCCATGACGCCAAGGAGCCCTCCATGAAGCGATCTCGTCAC TAAGTTCCCGTCTTCGTATGTGATTCATCTTTTGGATCATCCCCTCTTTTACTGGTATCCTTACACTTATCTATC CCCTGCCGTACTAATTCTATCTCATCCGTATATTCCTGACATCTAACATGTATCATCTTTGTACTGTATACTTAT ACCAAATGGAAGTTTGGTTGTATTTTTAACCCC |
| Length | 2958 |