CopciAB_456245
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_456245 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 456245 |
| Uniprot id | Functional description | Other 1 protein kinase | |
| Location | scaffold_4:150337..153162 | Strand | - |
| Gene length (nt) | 2826 | Transcript length (nt) | 2524 |
| CDS length (nt) | 2289 | Protein length (aa) | 762 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14858 | 24.9 | 1.704E-31 | 133 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_692 | 25.7 | 1.263E-19 | 94 |
| Lentinula edodes NBRC 111202 | Lenedo1_1157490 | 25.4 | 2.57E-19 | 93 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_195171 | 40.9 | 4.745E-6 | 49 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_456245.T0 |
| Description | Other 1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 556 | 659 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Other 1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
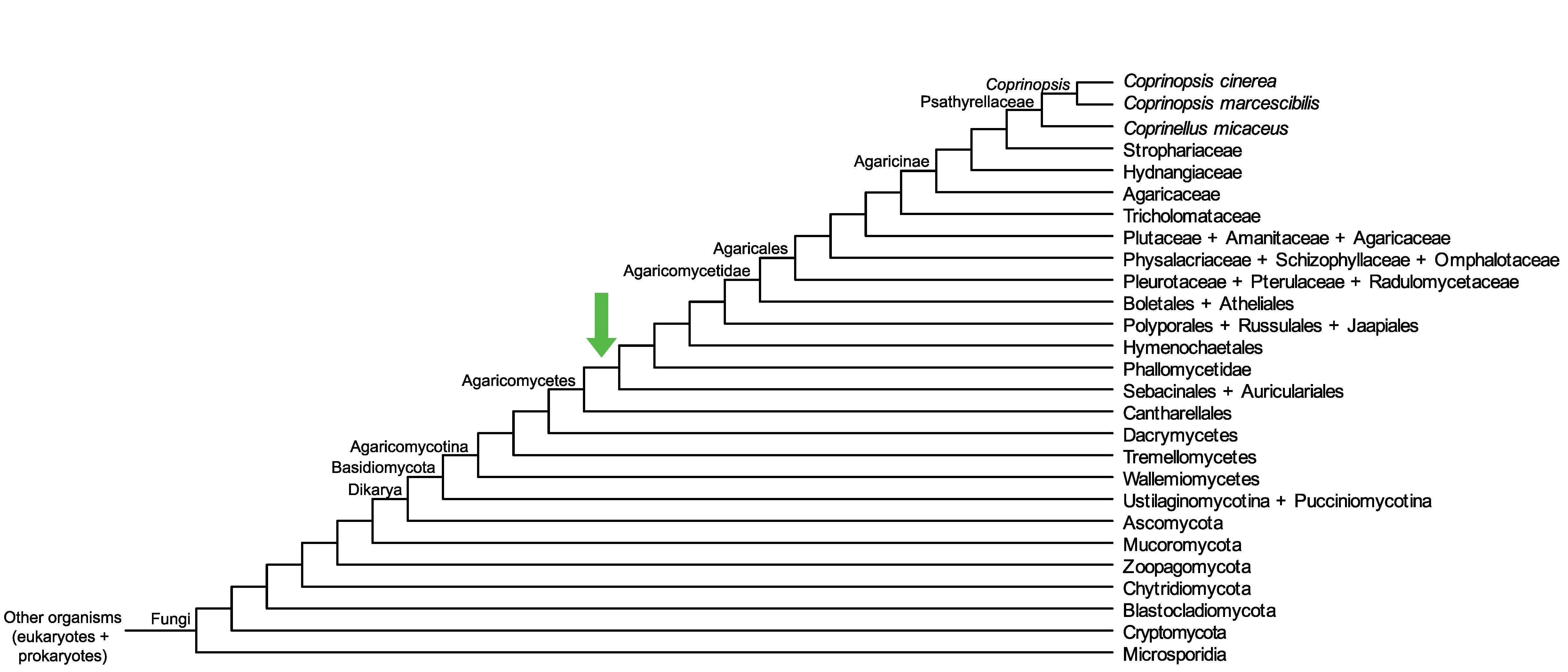
Conservation of CopciAB_456245 across fungi.
Arrow shows the origin of gene family containing CopciAB_456245.
Protein
| Sequence id | CopciAB_456245.T0 |
|---|---|
| Sequence |
>CopciAB_456245.T0 MTAETRTGSSIPQNQPVERPTCSSRPSNDASGTDVDSTMPVCDGRTRTTESVVGLLKQDLKDACYVGLRAMLTAF WGRMRAGVEGDEQFDAARAFDAFLTRAAGFCNDEASSGVKERIYEALKTRDQKESCAHLARALNLILEHGRGEDV AGFQHTVDDEETVVLVNNPTILKCHPIHDVYDYMPKRKPDLVRLPLSALAPIFPEREGERWEDWVESPFVKHGER VDWGSEGRRTLWRDVVDCWEVRKENVVSEDDLRLLVDRNYQFAIECALDTVDPTPHAAPSVPSPSAGQASGISCD SRKRKGHSDEHSDSDLESWKRVRTDYNSDSDSPHPTASPHAQSGLYALEMMRARWDRTHAIVVLLIDNQLSIQWY DPQGCIRTRPIDIVRQLPLLVATILVFQRFDNRMRGIGGFKLDVEVDGKAIPHTLGDDTRTRWGLTGRRSVGGQL PPCGDLASAVHENKIFGKFLWRAEKQESEEYILETAKRRSVHLGAYKEYVIDHLPCVKAYAEFGSFSTRHIREFM KLPSDGCLVPTGIAAEWLVPINDLPPEEYARAISQLIRCHFLLWQIGIAHGDISPSNLMQRRCPDGALQAVLNDF DLAAIMTPGDYSPNKAGFERTGTKPFMAIDLLCSPTKGRVNRMWYHDFESFLWCLVWYCSDVLRRNWSTGSFSKL AREKYWWLSYAPKRDSDDGVPPQKLDLWRTLLKFRDSWTGRHGIWDLAEEDWEKQLGPFRVFDKYFAKFDSCEDR GWLTFRGDSSMA |
| Length | 762 |
Coding
| Sequence id | CopciAB_456245.T0 |
|---|---|
| Sequence |
>CopciAB_456245.T0 ATGACGGCGGAGACTCGGACAGGGAGTTCTATCCCACAGAATCAACCCGTCGAACGACCGACGTGCAGCTCGCGC CCCTCAAACGATGCTTCGGGGACAGATGTAGATTCTACTATGCCTGTGTGTGATGGGAGAACGAGAACCACCGAG TCTGTGGTTGGTCTCCTCAAGCAGGATCTGAAAGACGCGTGCTATGTTGGTTTGCGGGCCATGTTAACTGCGTTT TGGGGGAGGATGCGTGCGGGCGTTGAAGGAGATGAGCAGTTTGATGCGGCGAGGGCGTTTGATGCGTTTCTGACT CGTGCTGCTGGGTTTTGCAATGACGAGGCGTCTAGCGGGGTTAAAGAAAGGATTTATGAAGCGTTGAAGACTCGA GACCAAAAGGAGAGTTGCGCCCATTTGGCTCGTGCGCTCAACTTGATCCTCGAGCATGGCCGAGGAGAGGACGTT GCGGGTTTTCAACACACCGTCGATGACGAGGAAACTGTCGTCCTCGTGAACAACCCCACGATTCTGAAGTGCCAT CCTATCCACGACGTTTATGACTACATGCCTAAACGGAAGCCTGACCTCGTCCGGTTGCCCTTGAGTGCGCTGGCG CCGATCTTCCCTGAGCGAGAAGGCGAGAGGTGGGAGGATTGGGTCGAGTCTCCGTTCGTGAAACATGGAGAGCGC GTTGATTGGGGATCGGAGGGACGGAGAACGCTTTGGCGTGATGTTGTGGATTGTTGGGAGGTTAGGAAGGAGAAT GTGGTGTCGGAGGATGATCTTAGGCTTCTTGTGGATCGAAACTATCAATTCGCGATTGAATGTGCTTTAGACACG GTTGACCCGACGCCGCATGCTGCTCCCTCGGTCCCATCGCCATCCGCTGGCCAGGCGTCAGGGATATCTTGTGAT AGCCGGAAACGGAAAGGACATTCAGACGAACACTCTGACAGTGACTTAGAATCGTGGAAGAGGGTCAGAACTGAC TACAACTCGGATTCGGACTCTCCCCATCCCACTGCATCCCCTCATGCTCAGTCAGGCTTGTATGCACTCGAGATG ATGCGAGCGAGATGGGACCGCACGCATGCTATCGTCGTCCTCTTGATCGATAATCAACTGTCCATCCAGTGGTAT GACCCACAAGGCTGTATCCGAACCCGGCCCATCGACATCGTCCGTCAGCTTCCTCTCCTCGTCGCGACGATCCTT GTTTTCCAACGATTTGATAATCGGATGCGGGGGATTGGAGGGTTCAAGCTGGATGTTGAAGTCGATGGGAAGGCG ATCCCGCACACGCTTGGAGACGACACGCGTACGCGGTGGGGGCTGACAGGGCGACGTTCAGTCGGTGGTCAGTTG CCCCCCTGTGGGGACCTAGCATCCGCTGTGCACGAGAACAAGATATTCGGCAAGTTTTTGTGGCGTGCGGAGAAG CAGGAGAGCGAAGAGTACATCCTCGAAACTGCCAAGAGGAGGTCGGTGCATCTCGGTGCGTACAAGGAGTATGTC ATCGACCACCTCCCCTGCGTCAAAGCCTACGCCGAGTTCGGCTCCTTCTCGACGAGGCATATTCGCGAGTTTATG AAGCTACCTTCGGATGGCTGCTTGGTGCCAACTGGAATCGCTGCTGAATGGTTGGTTCCGATCAACGACCTCCCA CCGGAGGAGTACGCTCGTGCGATCTCGCAACTCATTCGTTGTCATTTTCTTCTCTGGCAAATTGGTATCGCGCAT GGCGACATCAGTCCGTCGAACCTGATGCAGCGTCGTTGTCCGGATGGTGCCCTCCAAGCCGTCTTGAACGACTTT GATTTGGCTGCGATCATGACCCCAGGCGATTATTCACCCAATAAGGCGGGCTTCGAGCGGACGGGGACCAAGCCT TTCATGGCGATCGACCTTCTATGTTCCCCGACGAAAGGACGAGTGAATCGTATGTGGTATCACGACTTTGAGTCC TTCTTGTGGTGCTTGGTGTGGTACTGTTCCGATGTCCTGCGCCGCAATTGGTCCACTGGTTCATTTTCAAAGCTT GCGAGGGAAAAATACTGGTGGTTATCCTATGCTCCTAAGCGTGATAGTGACGATGGTGTACCGCCACAGAAACTG GACCTTTGGAGAACACTGTTGAAGTTCCGAGATAGTTGGACTGGCCGTCATGGGATTTGGGATCTAGCTGAGGAA GACTGGGAGAAGCAGCTGGGTCCATTCAGGGTGTTTGACAAGTACTTTGCGAAGTTTGATTCATGCGAGGATCGG GGTTGGTTGACGTTTCGGGGTGACTCCTCCATGGCTTAG |
| Length | 2289 |
Transcript
| Sequence id | CopciAB_456245.T0 |
|---|---|
| Sequence |
>CopciAB_456245.T0 GCGACCACCGCGCACTTACCGATCGAGAGAGGAGGAGTGCCTGGCAGGGGACGTAGCAAGTGGAGCGGGGAGCAT CTGAATCAGTCTGACCCTTTCCCCCGCTGCCGCGGCGAAGGTCGAAAGCGCCCGCTGCTGTTCCCCGACTGTGAC CACCACCAACACCTCGATGACGGCGGAGACTCGGACAGGGAGTTCTATCCCACAGAATCAACCCGTCGAACGACC GACGTGCAGCTCGCGCCCCTCAAACGATGCTTCGGGGACAGATGTAGATTCTACTATGCCTGTGTGTGATGGGAG AACGAGAACCACCGAGTCTGTGGTTGGTCTCCTCAAGCAGGATCTGAAAGACGCGTGCTATGTTGGTTTGCGGGC CATGTTAACTGCGTTTTGGGGGAGGATGCGTGCGGGCGTTGAAGGAGATGAGCAGTTTGATGCGGCGAGGGCGTT TGATGCGTTTCTGACTCGTGCTGCTGGGTTTTGCAATGACGAGGCGTCTAGCGGGGTTAAAGAAAGGATTTATGA AGCGTTGAAGACTCGAGACCAAAAGGAGAGTTGCGCCCATTTGGCTCGTGCGCTCAACTTGATCCTCGAGCATGG CCGAGGAGAGGACGTTGCGGGTTTTCAACACACCGTCGATGACGAGGAAACTGTCGTCCTCGTGAACAACCCCAC GATTCTGAAGTGCCATCCTATCCACGACGTTTATGACTACATGCCTAAACGGAAGCCTGACCTCGTCCGGTTGCC CTTGAGTGCGCTGGCGCCGATCTTCCCTGAGCGAGAAGGCGAGAGGTGGGAGGATTGGGTCGAGTCTCCGTTCGT GAAACATGGAGAGCGCGTTGATTGGGGATCGGAGGGACGGAGAACGCTTTGGCGTGATGTTGTGGATTGTTGGGA GGTTAGGAAGGAGAATGTGGTGTCGGAGGATGATCTTAGGCTTCTTGTGGATCGAAACTATCAATTCGCGATTGA ATGTGCTTTAGACACGGTTGACCCGACGCCGCATGCTGCTCCCTCGGTCCCATCGCCATCCGCTGGCCAGGCGTC AGGGATATCTTGTGATAGCCGGAAACGGAAAGGACATTCAGACGAACACTCTGACAGTGACTTAGAATCGTGGAA GAGGGTCAGAACTGACTACAACTCGGATTCGGACTCTCCCCATCCCACTGCATCCCCTCATGCTCAGTCAGGCTT GTATGCACTCGAGATGATGCGAGCGAGATGGGACCGCACGCATGCTATCGTCGTCCTCTTGATCGATAATCAACT GTCCATCCAGTGGTATGACCCACAAGGCTGTATCCGAACCCGGCCCATCGACATCGTCCGTCAGCTTCCTCTCCT CGTCGCGACGATCCTTGTTTTCCAACGATTTGATAATCGGATGCGGGGGATTGGAGGGTTCAAGCTGGATGTTGA AGTCGATGGGAAGGCGATCCCGCACACGCTTGGAGACGACACGCGTACGCGGTGGGGGCTGACAGGGCGACGTTC AGTCGGTGGTCAGTTGCCCCCCTGTGGGGACCTAGCATCCGCTGTGCACGAGAACAAGATATTCGGCAAGTTTTT GTGGCGTGCGGAGAAGCAGGAGAGCGAAGAGTACATCCTCGAAACTGCCAAGAGGAGGTCGGTGCATCTCGGTGC GTACAAGGAGTATGTCATCGACCACCTCCCCTGCGTCAAAGCCTACGCCGAGTTCGGCTCCTTCTCGACGAGGCA TATTCGCGAGTTTATGAAGCTACCTTCGGATGGCTGCTTGGTGCCAACTGGAATCGCTGCTGAATGGTTGGTTCC GATCAACGACCTCCCACCGGAGGAGTACGCTCGTGCGATCTCGCAACTCATTCGTTGTCATTTTCTTCTCTGGCA AATTGGTATCGCGCATGGCGACATCAGTCCGTCGAACCTGATGCAGCGTCGTTGTCCGGATGGTGCCCTCCAAGC CGTCTTGAACGACTTTGATTTGGCTGCGATCATGACCCCAGGCGATTATTCACCCAATAAGGCGGGCTTCGAGCG GACGGGGACCAAGCCTTTCATGGCGATCGACCTTCTATGTTCCCCGACGAAAGGACGAGTGAATCGTATGTGGTA TCACGACTTTGAGTCCTTCTTGTGGTGCTTGGTGTGGTACTGTTCCGATGTCCTGCGCCGCAATTGGTCCACTGG TTCATTTTCAAAGCTTGCGAGGGAAAAATACTGGTGGTTATCCTATGCTCCTAAGCGTGATAGTGACGATGGTGT ACCGCCACAGAAACTGGACCTTTGGAGAACACTGTTGAAGTTCCGAGATAGTTGGACTGGCCGTCATGGGATTTG GGATCTAGCTGAGGAAGACTGGGAGAAGCAGCTGGGTCCATTCAGGGTGTTTGACAAGTACTTTGCGAAGTTTGA TTCATGCGAGGATCGGGGTTGGTTGACGTTTCGGGGTGACTCCTCCATGGCTTAGTGTAGGGCATGTGTGTCCTT ACGAGTACGTAGAATTTGAAGATGCCTCCCTAAGGCCATTGACTATTTC |
| Length | 2524 |
Gene
| Sequence id | CopciAB_456245.T0 |
|---|---|
| Sequence |
>CopciAB_456245.T0 GCGACCACCGCGCACTTACCGATCGAGAGAGGAGGAGTGCCTGGCAGGGGACGTAGCAAGTGGAGCGGGGAGCAT CTGAATCAGTCTGACCCTTTCCCCCGCTGCCGCGGCGAAGGTCGAAAGCGCCCGCTGCTGTTCCCCGACTGTGAC CACCACCAACACCTCGATGACGGCGGAGACTCGGACAGGGAGTTCTATCCCACAGAATCAACCCGTCGAACGACC GACGTGCAGCTCGCGCCCCTCAAACGATGCTTCGGGGACAGATGTAGATTCTACTATGCCTGTGTGTGATGGGAG GTTCGTGTCGTTTATTCTTCTTCAGCGAGCTCAGAACGTTGATTACTGTCTGTCTATTTGAATAGAACGAGAACC ACCGAGTCTGTGGTTGGTCTCCTCAAGCAGGATCTGAAAGACGCGTGCTATGTTGGTTTGCGGGCCATGTTAACT GCGTTTTGGGGGAGGATGCGTGCGGGCGTTGAAGGAGATGAGCAGTTTGATGCGGCGAGGGCGTTTGATGCGTTT CTGACTCGTGCTGCTGGGTTTTGCAATGACGAGGCGTCTAGCGGGGTTAAAGAAAGGATTTATGAAGCGTATGTT GACTGGCTTAGATTTGCTCTCCTATCTCTATGAGACTGACTAGCATCCTTCGCTTTTTCAGGTTGAAGACTCGAG ACCAAAAGGAGAGTTGCGCCCATTTGGCTCGTGCGCTCAACTTGATCCTCGAGCATGGCCGAGGAGAGGACGTTG CGGGTTTTCAACACACCGTCGATGACGAGGAAACTGTCGTCCTCGTGAACAACCCCACGATTCTGAAGTGCCATC CTATCCACGACGTTTATGACTACATGCCTAAACGGAAGCCTGACCTCGTCCGGTTGCCCTTGAGTGCGCTGGCGC CGATCTTCCCTGAGCGAGAAGGCGAGAGGTGGGAGGATTGGGTCGAGTCTCCGTTCGTGAAACATGGAGAGCGCG TTGATTGGGGATCGGAGGGACGGAGAACGCTTTGGCGTGATGTTGTGGATTGTTGGGAGGTTAGGAAGGAGAATG TGGTGTCGGAGGATGATCTTAGGCTTCTTGTGGATCGAAACTATCAATTCGCGATTGAATGTGGTGGATACATGT CTCTTGGTCTGCACCGGGCCGCATTGAACTGATCGATTTTCGTTTGGATTAGCTTTAGACACGGTTGACCCGACG CCGCATGCTGCTCCCTCGGTCCCATCGCCATCCGCTGGCCAGGCGTCAGGGATATCTTGTGATAGCCGGAAACGG AAAGGACATTCAGACGAACACTCTGACAGTGACTTAGAATCGTGGAAGAGGGTCAGAACTGACTACAACTCGGAT TCGGACTCTCCCCATCCCACTGCATCCCCTCATGCTCAGTCAGGCTTGTATGCACTCGAGATGATGCGAGCGAGA TGGGACCGCACGCATGCTATCGTCGTCCTCTTGATCGGTAGGTGCCAAAATCCTTTAGTCTTCCTTCCAAATCTG ACTGTTTTGTAGATAATCAACTGTCCATCCAGTGGTATGACCCACAAGGCTGTATCCGAACCCGGCCCATCGACA TCGTCCGTCAGCTTCCTCTCCTCGTCGCGACGATCCTTGTTTTCCAACGATTTGATAATCGGATGCGGGGGATTG GAGGGTTCAAGCTGGATGTTGAAGTCGATGGGAAGGCGATCCCGCACACGCTTGGAGACGACACGCGTACGCGGT GGGGGCTGACAGGGCGACGTTCAGTCGGTGGTCAGTTGCCCCCCTGTGGGGACCTAGCATCCGCTGTGCACGAGA ACAAGATATTCGGCAAGTTTTTGTGGCGTGCGGAGAAGCAGGAGAGCGAAGAGTACATCCTCGAAACTGCCAAGA GGAGGTCGGTGCATCTCGGTGCGTACAAGGAGTATGTCATCGACCACCTCCCCTGCGTCAAAGCCTACGCCGAGT TCGGCTCCTTCTCGACGAGGCATATTCGCGAGTTTATGAAGCTACCTTCGGATGGCTGCTTGGTGCCAACTGGAA TCGCTGCTGAATGGTTGGTTCCGATCAACGACCTCCCACCGGAGGAGTACGCTCGTGCGATCTCGCAACTCATTC GTTGTGAGTACGGGTTTTCTGCCTAGTTTACCCATCGAGCTAAGTCCCTTTTCATTAGGTCATTTTCTTCTCTGG CAAATTGGTATCGCGCATGGCGACATCAGTCCGTCGAACCTGATGCAGCGTCGTTGTCCGGATGGTGCCCTCCAA GCCGTCTTGAACGACTTTGATTTGGCTGCGATCATGACCCCAGGCGATTATTCACCCAATAAGGCGGGCTTCGAG CGGACGGGGACCAAGCCTTTCATGGCGATCGACCTTCTATGTTCCCCGACGAAAGGACGAGTGAATCGTATGTGG TATCACGACTTTGAGTCCTTCTTGTGGTGCTTGGTGTGGTACTGTTCCGATGTCCTGCGCCGCAATTGGTCCACT GGTTCATTTTCAAAGCTTGCGAGGGAAAAATACTGGTGGTTATCCTATGCTCCTAAGCGTGATAGTGACGATGGT GTACCGCCACAGAAACTGGACCTTTGGAGAACACTGTTGAAGTTCCGAGATAGTTGGACTGGCCGTCATGGGATT TGGGATCTAGCTGAGGAAGACTGGGAGAAGCAGCTGGGTCCATTCAGGGTGTTTGACAAGTACTTTGCGAAGTTT GATTCATGCGAGGATCGGGGTTGGTTGACGTTTCGGGGTGACTCCTCCATGGCTTAGTGTAGGGCATGTGTGTCC TTACGAGTACGTAGAATTTGAAGATGCCTCCCTAAGGCCATTGACTATTTC |
| Length | 2826 |