CopciAB_460475
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_460475 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | SSN3 | Synonyms | 460475 |
| Uniprot id | Functional description | Serine/Threonine protein kinases, catalytic domain | |
| Location | scaffold_5:441118..443083 | Strand | + |
| Gene length (nt) | 1966 | Transcript length (nt) | 1552 |
| CDS length (nt) | 1347 | Protein length (aa) | 448 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | stk-8 |
| Cryptococcus neoformans | SSN3 |
| Saccharomyces cerevisiae | YPL042C_SSN3 |
| Schizosaccharomyces pombe | srb10 |
| Ustilago maydis | SSN3 |
| Aspergillus nidulans | ssn3 |
| Candida albicans | SSN3 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7259444 | 79.7 | 4.568E-234 | 720 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB20045 | 79 | 5.627E-233 | 717 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_212134 | 74.6 | 5.545E-228 | 702 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_56945 | 74.6 | 5.779E-228 | 702 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1093132 | 75.6 | 1.571E-217 | 672 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1385059 | 75.1 | 2.02E-216 | 669 |
| Flammulina velutipes | Flave_chr11AA01178 | 77.3 | 2.212E-214 | 663 |
| Pleurotus ostreatus PC9 | PleosPC9_1_114610 | 75.5 | 1.544E-208 | 646 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_114554 | 71.1 | 2.632E-203 | 631 |
| Schizophyllum commune H4-8 | Schco3_58330 | 82.1 | 1.462E-194 | 606 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_8367 | 70.8 | 3.097E-193 | 602 |
| Lentinula edodes NBRC 111202 | Lenedo1_1054076 | 75.7 | 4.431E-189 | 590 |
| Auricularia subglabra | Aurde3_1_1219939 | 74.7 | 4.588E-185 | 579 |
| Lentinula edodes B17 | Lened_B_1_1_1408 | 55.3 | 3.049E-144 | 460 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | SSN3 |
|---|---|
| Protein id | CopciAB_460475.T0 |
| Description | Serine/Threonine protein kinases, catalytic domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd07842 | STKc_CDK8_like | - | 44 | 352 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 49 | 352 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K02208 |
EggNOG
| COG category | Description |
|---|---|
| K | Serine/Threonine protein kinases, catalytic domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
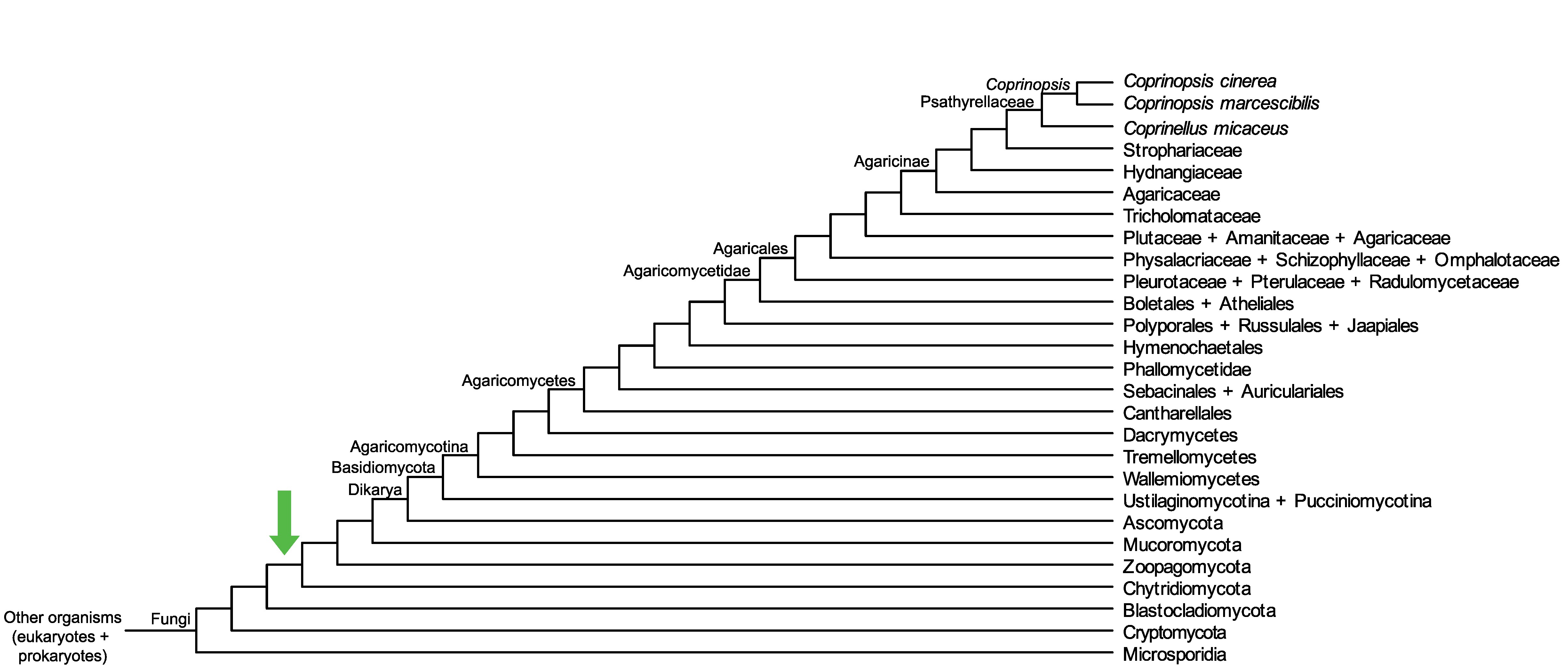
Conservation of CopciAB_460475 across fungi.
Arrow shows the origin of gene family containing CopciAB_460475.
Protein
| Sequence id | CopciAB_460475.T0 |
|---|---|
| Sequence |
>CopciAB_460475.T0 MQQQPQQQQQQVSLDEWVSNVMQHDPMRVYRARRDAHHRPVTSKYAILGFISSGTYGRVYKAQSINGGELLAIKK FKPDKEGDVVTYTGISQSAIREIALNREINHENVVALREVILEDKSIYMVFEYAEHDFLQVIHHYYQTIRTSIPT AVLKSLIYQLFNGLIYLHASHILHRDLKPANILITSQGVVKIGDLGLARLCYEPLQPLFAGDKVVVTIWYRAPEL LMGAKHYNKAIDCWAVGCVMAELASLRPIFKGEEAKLDSKKNVPFQRDQLIKIFEVLGTPDERDWPGVVDMPEYR NMKLLDHFSNRLSDWCHSRIRSPQGYDLLRQLFAYDPDNRLTADQAVQHKWFHEDPLPTWNAFQSLAQHQIPPCR RITQDEAPSMMPMATQQNTQQAMGGGHIAGAGAPFSKPGSTASFASLSGGGPYAQSQSGTGGHTHTRKKARMG |
| Length | 448 |
Coding
| Sequence id | CopciAB_460475.T0 |
|---|---|
| Sequence |
>CopciAB_460475.T0 ATGCAGCAACAACCACAACAGCAACAACAACAAGTATCATTGGACGAATGGGTTTCCAATGTCATGCAACATGAT CCCATGCGTGTCTACCGCGCTAGACGGGATGCGCACCATCGACCGGTTACCTCGAAATATGCGATCCTAGGCTTC ATTTCTTCTGGAACGTATGGACGCGTGTATAAAGCTCAGTCTATCAACGGAGGAGAGCTTCTGGCGATTAAAAAG TTCAAACCAGACAAAGAGGGCGATGTGGTCACCTACACTGGAATCAGCCAGAGTGCAATTCGTGAAATCGCGCTG AATAGAGAAATAAACCACGAGAATGTAGTGGCCCTCCGAGAAGTAATCCTCGAGGATAAATCCATCTACATGGTC TTCGAATACGCAGAACACGACTTTCTCCAAGTAATACACCACTACTACCAAACCATCCGCACCTCAATACCAACC GCAGTCCTCAAATCCCTAATCTACCAACTCTTCAACGGGCTCATCTACCTCCACGCCTCCCACATCCTCCACCGC GACCTCAAACCAGCAAACATCCTCATCACCTCCCAAGGCGTCGTCAAAATCGGCGACCTGGGTCTCGCACGTCTG TGTTACGAGCCTCTCCAACCGCTCTTCGCCGGCGACAAGGTCGTCGTAACGATCTGGTACCGCGCACCGGAACTC TTGATGGGCGCGAAGCACTACAATAAAGCGATAGATTGCTGGGCTGTGGGGTGTGTTATGGCTGAGTTGGCGAGC CTGAGGCCGATTTTTAAAGGTGAAGAGGCGAAGCTGGACTCGAAGAAGAATGTACCGTTTCAGAGGGACCAGTTG ATCAAGATTTTCGAGGTGCTGGGAACGCCGGATGAACGAGACTGGCCAGGTGTAGTCGACATGCCTGAATATCGA AATATGAAACTCCTGGACCACTTCTCAAACCGCCTATCAGATTGGTGCCACTCCCGTATACGGTCACCTCAAGGC TACGACCTCCTCCGCCAGCTCTTCGCCTACGATCCCGATAACAGACTAACAGCAGACCAGGCTGTTCAGCATAAA TGGTTTCACGAGGATCCTTTGCCTACCTGGAACGCCTTCCAATCCCTCGCACAACACCAAATCCCGCCATGCCGA CGTATAACCCAAGACGAGGCGCCATCCATGATGCCCATGGCCACCCAGCAAAACACGCAACAAGCAATGGGGGGA GGACACATTGCAGGAGCAGGCGCGCCATTCTCCAAACCGGGTAGTACAGCCAGTTTTGCAAGCCTGAGCGGTGGT GGGCCGTACGCCCAGAGTCAGAGTGGGACCGGAGGTCACACGCATACGAGGAAGAAGGCTAGAATGGGGTAG |
| Length | 1347 |
Transcript
| Sequence id | CopciAB_460475.T0 |
|---|---|
| Sequence |
>CopciAB_460475.T0 ATCTTCCTCCCACCGTCAACAACGACGATGAGCATCCTCATCGTCGCCTTCCATGCAGCAACAACCACAACAGCA ACAACAACAAGTATCATTGGACGAATGGGTTTCCAATGTCATGCAACATGATCCCATGCGTGTCTACCGCGCTAG ACGGGATGCGCACCATCGACCGGTTACCTCGAAATATGCGATCCTAGGCTTCATTTCTTCTGGAACGTATGGACG CGTGTATAAAGCTCAGTCTATCAACGGAGGAGAGCTTCTGGCGATTAAAAAGTTCAAACCAGACAAAGAGGGCGA TGTGGTCACCTACACTGGAATCAGCCAGAGTGCAATTCGTGAAATCGCGCTGAATAGAGAAATAAACCACGAGAA TGTAGTGGCCCTCCGAGAAGTAATCCTCGAGGATAAATCCATCTACATGGTCTTCGAATACGCAGAACACGACTT TCTCCAAGTAATACACCACTACTACCAAACCATCCGCACCTCAATACCAACCGCAGTCCTCAAATCCCTAATCTA CCAACTCTTCAACGGGCTCATCTACCTCCACGCCTCCCACATCCTCCACCGCGACCTCAAACCAGCAAACATCCT CATCACCTCCCAAGGCGTCGTCAAAATCGGCGACCTGGGTCTCGCACGTCTGTGTTACGAGCCTCTCCAACCGCT CTTCGCCGGCGACAAGGTCGTCGTAACGATCTGGTACCGCGCACCGGAACTCTTGATGGGCGCGAAGCACTACAA TAAAGCGATAGATTGCTGGGCTGTGGGGTGTGTTATGGCTGAGTTGGCGAGCCTGAGGCCGATTTTTAAAGGTGA AGAGGCGAAGCTGGACTCGAAGAAGAATGTACCGTTTCAGAGGGACCAGTTGATCAAGATTTTCGAGGTGCTGGG AACGCCGGATGAACGAGACTGGCCAGGTGTAGTCGACATGCCTGAATATCGAAATATGAAACTCCTGGACCACTT CTCAAACCGCCTATCAGATTGGTGCCACTCCCGTATACGGTCACCTCAAGGCTACGACCTCCTCCGCCAGCTCTT CGCCTACGATCCCGATAACAGACTAACAGCAGACCAGGCTGTTCAGCATAAATGGTTTCACGAGGATCCTTTGCC TACCTGGAACGCCTTCCAATCCCTCGCACAACACCAAATCCCGCCATGCCGACGTATAACCCAAGACGAGGCGCC ATCCATGATGCCCATGGCCACCCAGCAAAACACGCAACAAGCAATGGGGGGAGGACACATTGCAGGAGCAGGCGC GCCATTCTCCAAACCGGGTAGTACAGCCAGTTTTGCAAGCCTGAGCGGTGGTGGGCCGTACGCCCAGAGTCAGAG TGGGACCGGAGGTCACACGCATACGAGGAAGAAGGCTAGAATGGGGTAGAATGAAAGTTGAATTTACAGACTTAT GGCCCTTAGGTGCTACACACTAGGACTACTACAAACAAGGGCTACAGCAATACATCAGAACCACCTCCACTACCA CCCATGTACATCGCATAGAAAGGCGGCTGGTTAATCCTAAACTCAAACCTCC |
| Length | 1552 |
Gene
| Sequence id | CopciAB_460475.T0 |
|---|---|
| Sequence |
>CopciAB_460475.T0 ATCTTCCTCCCACCGTCAACAACGACGATGAGCATCCTCATCGTCGCCTTCCATGCAGCAACAACCACAACAGCA ACAACAACAAGTATCATTGGACGAATGGGTTTCCAATGTCATGCAACATGATCCCATGCGTGTCTACCGCGCTAG ACGGGATGCGCACCATCGACCGGTTACCTCGAAATATGCGATCCTAGGCTTCATTTCTTCTGGAACGTATGGACG CGTGTATAAAGCTCAGTCTATCAACGGAGGAGAGCTTCTGGCGATTAAAAAGTTCAAACCAGACAAAGAGGGCGA TGTGGTCACCTACACTGGAATCAGCCAGAGTGCAATTCGTGAAATCGCGGTGCGTTTTTTCTCGCACTCTTCTTT GAGAGCTTTGAATTGACTATTATCTAGCTGAATAGAGAAATAAACCACGAGAATGTAGTGGCCCTCCGAGAAGTA ATCCTCGAGGATAAATCCATCTACATGGTCTTCGAATACGCAGAACACGACTTTCTCGTTCGTCCCTCTTGTCCT TCTTCCAATCACAACTCTAACCGACTCTGATTACAGCAAGTAATACACCACTACTACCAAACCATCCGCACCTCA ATACCAACCGCAGTCCTCAAATCCCTAATCTACCAACTCTTCAACGGGCTCATCTACCTCCACGCCTCCCACATC CTCCACCGCGACCTCAAACCAGCAAACATCCTCATCACCTCCCAAGGCGTCGTCAAAATCGGCGACCTGGGTCTC GCACGTCTGTGTTACGAGCCTCTCCAACCGCTCTTCGCCGGCGACAAGGTCGTCGTAACGATCTGGTACCGCGCA CCGGAACTCTTGATGGGCGCGAAGCACTACAATAAAGCGATAGATTGCTGGGCTGTGGGGTGTGTTATGGCTGAG TTGGCGAGCCTGAGGCCGATTTTTAAAGGTGAAGAGGCGAAGCTGGACTCGAAGAAGAATGTACCGTTTCAGAGG GACCAGTTGATCAAGATTTTCGAGGTGCTGGGAACGCCGGATGGTGCGTCGATCGCTTTCTCTCCTCCCCTCCTC TCTGACAGGCGTTTCTTTTACTCTCAGAACGAGACTGGCCAGGTGTAGTCGACATGCCTGAATATCGAAATATGA AACTCCTGGACCAGTGAGCTCACGACCCCCCTCCCCTCCTTAAACCCCATTCTGACTTTCACAAACCAGCTTCTC AAACCGCCTATCAGATTGGTGCCACTCCCGTATACGGTCACCTCAAGGCTACGACCTCCTCCGCCAGCTCTTCGC CTACGATCCCGATAACAGACTAACAGCAGACCAGGCTGTTCAGCATAAATGGTTTCACGAGGATCCTTTGCCTAC CTGGAAGTACGTCTCTCAACCCCGTATCTAAGTTGTTTCGTTCTACGACATTGCTCATCCTCTTTCTCCGATAGC GCCTTCCAATCCCTCGCACAACACCAAATCCCGCCATGCCGACGTATAACCCAAGACGAGGCGCCATCCATGATG CCCATGGCCACCCAGCAAAACACGCAACAAGCAATGGGGGGAGGACACATTGCAGGTATGCACTCCTAATCGCGT ATGGACTACGGGCGAGGACCTAGCTATGAAAACGTCTTTCCCCATTGCACAGGAGCAGGCGCGCCATTCTCCAAA CCGGGTAGTACAGCCAGTTTTGCAAGCCTGAGCGGTGGTGGGCCGTACGCCCAGAGTCAGAGTGGGACCGGAGGT CACACGCATACGAGGAAGAAGGCTAGAATGGGGTAGAATGAAAGTTGAATTTACAGTAAGGTCTTGCATAATCTT CCAATAGTCGCCAATTACTAACTACGCACCAGGACTTATGGCCCTTAGGTGCTACACACTAGGACTACTACAAAC AAGGGCTACAGCAATACATCAGAACCACCTCCACTACCACCCATGTACATCGCATAGAAAGGCGGCTGGTTAATC CTAAACTCAAACCTCC |
| Length | 1966 |