CopciAB_461315
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_461315 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 461315 |
| Uniprot id | Functional description | ABC1 family | |
| Location | scaffold_10:852461..855542 | Strand | + |
| Gene length (nt) | 3082 | Transcript length (nt) | 2599 |
| CDS length (nt) | 2334 | Protein length (aa) | 777 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YGL119W_COQ8 |
| Neurospora crassa | stk-25 |
| Aspergillus nidulans | AN6572 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264059 | 50.5 | 1.207E-215 | 684 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB23565 | 62.9 | 2.627E-206 | 657 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_76847 | 65 | 1.85E-205 | 654 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_122165 | 57.9 | 3.919E-205 | 653 |
| Grifola frondosa | Grifr_OBZ74946 | 59.8 | 4.801E-194 | 621 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1048694 | 61.3 | 5.155E-192 | 615 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87723 | 66.6 | 2.627E-190 | 610 |
| Flammulina velutipes | Flave_chr01AA00398 | 56.9 | 9.579E-184 | 591 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1378577 | 65.7 | 5.075E-183 | 589 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_42159 | 63.6 | 6.433E-178 | 574 |
| Schizophyllum commune H4-8 | Schco3_1171804 | 62.1 | 5.32E-172 | 557 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4936 | 42.8 | 6.768E-166 | 539 |
| Lentinula edodes NBRC 111202 | Lenedo1_1041859 | 50.1 | 4.727E-159 | 519 |
| Auricularia subglabra | Aurde3_1_1159159 | 51.3 | 2.651E-150 | 494 |
| Lentinula edodes B17 | Lened_B_1_1_9226 | 38.7 | 4.261E-88 | 309 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_461315.T0 |
| Description | ABC1 family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd13970 | ABC1_ADCK3 | IPR034646 | 368 | 659 |
| Pfam | PF03109 | ABC1 atypical kinase-like domain | IPR004147 | 372 | 652 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR034646 | ADCK3-like domain |
| IPR004147 | ABC1 atypical kinase-like domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| K08869 |
EggNOG
| COG category | Description |
|---|---|
| S | ABC1 family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
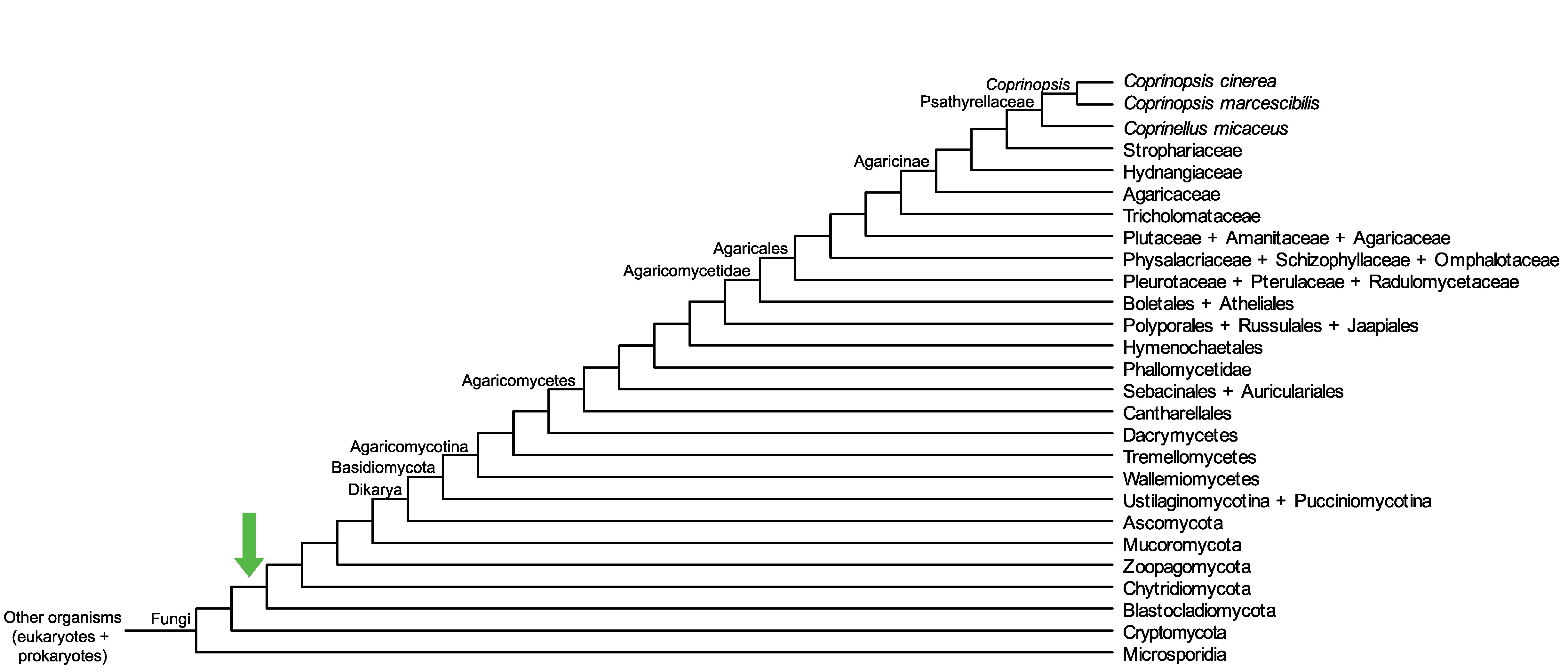
Conservation of CopciAB_461315 across fungi.
Arrow shows the origin of gene family containing CopciAB_461315.
Protein
| Sequence id | CopciAB_461315.T0 |
|---|---|
| Sequence |
>CopciAB_461315.T0 MPPGPAYNALAVVYSVARIVEHAARHRAAAGARTLGVSEDILTTTRPGPAPQPAPTAPTSSTPSTSTSSDASAST SPSEERVGEASTQTQPKSQLSKAERRRIQAQLEEEEFRAILGKRWVDGVLVDGSLPFSVSHSHPAQIISEAPRDP VPPIPEIPELPTQSQSPAESLESGTKTWVEKEREAEEVARVVEEVSHEIEHERELETKERPIPQVAQKYQSSHQQ ASSSSATSVPTSNPTAAIEASDPSPTQAASTSPSDLSDSPSCPLPVTTPTPPPKKVLQASKVPSSRIGRLFHYGG LAASLGYGAAAELLRGSDSSNATGNVMLSEANIKRLVGKLTQMRGAALKLGQFMSIQDTHLLPPELDQIFRRVQD SAHYMPDWQMEKVLSAAYGPNWCNPSNPEETVFMEFERIPFAAASIGQVHRARLAPRLCPPELLDQGNKDVGLDV AVKVQFPNIVNSIKSDLGYVKMLLSVGGLLPKGLFLDRTIEVMSAELADECSYTREASFLKKFRAKKFLGGDERF KVPWVWEGSTDQVLVMERVGGVGVGDVSHGSALEEGEVAVDEETVRGLSQRDRNDIAARIIELCLKELFEFRAMQ TDPNWTNFLWNQKTRQIELVDFGATRTYSKKFMDSWLRLLQAAASEDRQTCIDESIKIGYLTGQEDEVMLDAHVQ SMTLLATPFKETTTQPFAFGPGSRWSEVTKEIREFIPVMVKRRLTPPPKETYSLNRKLSGAFLLAARLDAKVDTK EIWDRIVGKYEFGRVEEEAAELAKDID |
| Length | 777 |
Coding
| Sequence id | CopciAB_461315.T0 |
|---|---|
| Sequence |
>CopciAB_461315.T0 ATGCCGCCAGGCCCGGCGTACAATGCTCTCGCTGTGGTCTATTCTGTCGCTCGGATCGTCGAGCATGCGGCACGA CATCGAGCTGCAGCTGGCGCTCGAACTTTGGGCGTCTCAGAGGATATACTGACGACGACACGGCCTGGACCAGCA CCACAACCCGCACCAACTGCACCAACATCCTCGACCCCTTCGACTTCAACCTCGTCCGACGCTTCAGCTAGCACA AGCCCGTCCGAGGAGAGGGTAGGAGAAGCCTCTACACAGACTCAACCAAAATCTCAGCTCTCCAAAGCAGAACGA CGACGGATACAAGCGCAACTAGAAGAGGAAGAGTTCCGAGCGATACTTGGAAAACGATGGGTTGATGGCGTATTG GTCGACGGGTCTCTCCCATTCAGCGTTTCGCACAGCCATCCAGCTCAAATTATATCTGAAGCACCCAGGGACCCT GTGCCGCCGATACCAGAAATACCAGAACTGCCAACTCAGTCGCAGTCGCCGGCAGAGTCTTTGGAGTCGGGGACG AAGACGTGGGTGGAGAAGGAAAGAGAGGCAGAGGAAGTTGCGAGAGTTGTGGAGGAGGTTAGCCATGAAATCGAA CACGAACGCGAATTGGAAACGAAGGAAAGGCCTATACCTCAGGTGGCGCAGAAGTACCAGTCCAGTCACCAGCAG GCATCGTCCTCATCTGCAACCAGTGTTCCCACTTCAAACCCGACTGCTGCAATCGAAGCTTCTGACCCTTCACCG ACACAGGCAGCTTCTACGTCGCCTTCAGACCTTTCAGACTCTCCCTCATGCCCTCTTCCCGTCACAACCCCTACA CCACCACCCAAAAAGGTGCTTCAAGCATCTAAAGTTCCCTCGTCACGGATAGGCAGATTATTCCACTATGGAGGT CTCGCTGCTTCACTAGGCTATGGAGCCGCCGCTGAACTCCTTAGAGGGAGTGATAGCTCCAACGCAACAGGCAAC GTCATGCTCTCAGAAGCGAACATCAAGCGATTGGTGGGCAAGCTTACCCAGATGCGAGGGGCCGCTCTCAAGCTT GGGCAGTTCATGAGTATCCAAGATACACATCTCCTCCCACCAGAGCTCGACCAGATATTCAGACGTGTCCAGGAT AGCGCGCATTACATGCCAGATTGGCAAATGGAGAAAGTCCTCTCAGCGGCGTACGGTCCTAACTGGTGCAACCCT TCAAATCCCGAAGAAACTGTCTTCATGGAGTTCGAGCGGATACCGTTCGCGGCTGCGTCCATCGGTCAGGTGCAC CGAGCCAGGCTAGCACCGCGACTTTGCCCACCCGAGTTGCTCGACCAGGGTAACAAGGATGTTGGCCTAGACGTC GCAGTCAAAGTCCAATTCCCAAACATCGTAAACTCGATAAAGAGTGACTTGGGTTATGTCAAAATGCTACTCAGC GTCGGAGGTCTGCTGCCTAAAGGTCTTTTCCTGGACAGGACGATCGAGGTGATGTCCGCGGAACTAGCAGACGAA TGCTCCTACACCCGCGAAGCCTCGTTCCTCAAGAAATTCAGGGCGAAAAAATTCTTGGGTGGAGACGAGCGGTTT AAAGTACCTTGGGTGTGGGAGGGTAGCACCGATCAGGTGTTGGTCATGGAGCGGGTAGGCGGGGTGGGTGTTGGG GATGTCAGTCATGGTAGTGCTCTCGAGGAAGGCGAGGTGGCGGTTGACGAGGAGACTGTGAGGGGTTTGAGCCAG AGGGATCGGAATGACATTGCTGCTCGAATCATTGAACTGTGCCTGAAAGAGCTCTTCGAGTTCCGGGCGATGCAG ACAGATCCCAATTGGACAAACTTCCTCTGGAACCAAAAGACCAGGCAGATCGAACTCGTCGACTTTGGCGCAACA CGCACATACAGCAAAAAGTTCATGGACAGCTGGTTAAGGCTGCTCCAAGCAGCCGCCTCCGAGGACCGCCAAACA TGTATCGATGAAAGCATCAAAATCGGATACTTGACGGGCCAAGAGGACGAAGTCATGCTCGATGCACATGTTCAG TCTATGACTCTACTCGCCACTCCTTTCAAAGAAACCACTACCCAGCCGTTTGCGTTCGGACCTGGCTCGAGGTGG TCGGAAGTGACGAAGGAGATTAGGGAGTTCATTCCTGTGATGGTGAAGAGGAGGTTGACCCCCCCGCCGAAGGAG ACATATAGTTTGAATAGAAAATTGAGCGGGGCGTTCCTCCTCGCTGCTAGACTCGATGCCAAGGTTGATACGAAG GAGATTTGGGATAGGATTGTTGGGAAGTACGAGTTTGGGCGTGTGGAAGAGGAGGCGGCGGAGTTGGCTAAAGAT ATTGATTGA |
| Length | 2334 |
Transcript
| Sequence id | CopciAB_461315.T0 |
|---|---|
| Sequence |
>CopciAB_461315.T0 GTGAATCTTTTGGTGACGACACCGACAACCGCGAAGACGGACGCGAGCTCATCGCGCTGCACGACGACAACGGGT CTCTTTTTCGTGTTCCTTATCGCGGCTATTGAGCTCTAGGACATCGGGCAACCAGGTCAACGTAAAGAAAGAAAG GCATGCCGCCAGGCCCGGCGTACAATGCTCTCGCTGTGGTCTATTCTGTCGCTCGGATCGTCGAGCATGCGGCAC GACATCGAGCTGCAGCTGGCGCTCGAACTTTGGGCGTCTCAGAGGATATACTGACGACGACACGGCCTGGACCAG CACCACAACCCGCACCAACTGCACCAACATCCTCGACCCCTTCGACTTCAACCTCGTCCGACGCTTCAGCTAGCA CAAGCCCGTCCGAGGAGAGGGTAGGAGAAGCCTCTACACAGACTCAACCAAAATCTCAGCTCTCCAAAGCAGAAC GACGACGGATACAAGCGCAACTAGAAGAGGAAGAGTTCCGAGCGATACTTGGAAAACGATGGGTTGATGGCGTAT TGGTCGACGGGTCTCTCCCATTCAGCGTTTCGCACAGCCATCCAGCTCAAATTATATCTGAAGCACCCAGGGACC CTGTGCCGCCGATACCAGAAATACCAGAACTGCCAACTCAGTCGCAGTCGCCGGCAGAGTCTTTGGAGTCGGGGA CGAAGACGTGGGTGGAGAAGGAAAGAGAGGCAGAGGAAGTTGCGAGAGTTGTGGAGGAGGTTAGCCATGAAATCG AACACGAACGCGAATTGGAAACGAAGGAAAGGCCTATACCTCAGGTGGCGCAGAAGTACCAGTCCAGTCACCAGC AGGCATCGTCCTCATCTGCAACCAGTGTTCCCACTTCAAACCCGACTGCTGCAATCGAAGCTTCTGACCCTTCAC CGACACAGGCAGCTTCTACGTCGCCTTCAGACCTTTCAGACTCTCCCTCATGCCCTCTTCCCGTCACAACCCCTA CACCACCACCCAAAAAGGTGCTTCAAGCATCTAAAGTTCCCTCGTCACGGATAGGCAGATTATTCCACTATGGAG GTCTCGCTGCTTCACTAGGCTATGGAGCCGCCGCTGAACTCCTTAGAGGGAGTGATAGCTCCAACGCAACAGGCA ACGTCATGCTCTCAGAAGCGAACATCAAGCGATTGGTGGGCAAGCTTACCCAGATGCGAGGGGCCGCTCTCAAGC TTGGGCAGTTCATGAGTATCCAAGATACACATCTCCTCCCACCAGAGCTCGACCAGATATTCAGACGTGTCCAGG ATAGCGCGCATTACATGCCAGATTGGCAAATGGAGAAAGTCCTCTCAGCGGCGTACGGTCCTAACTGGTGCAACC CTTCAAATCCCGAAGAAACTGTCTTCATGGAGTTCGAGCGGATACCGTTCGCGGCTGCGTCCATCGGTCAGGTGC ACCGAGCCAGGCTAGCACCGCGACTTTGCCCACCCGAGTTGCTCGACCAGGGTAACAAGGATGTTGGCCTAGACG TCGCAGTCAAAGTCCAATTCCCAAACATCGTAAACTCGATAAAGAGTGACTTGGGTTATGTCAAAATGCTACTCA GCGTCGGAGGTCTGCTGCCTAAAGGTCTTTTCCTGGACAGGACGATCGAGGTGATGTCCGCGGAACTAGCAGACG AATGCTCCTACACCCGCGAAGCCTCGTTCCTCAAGAAATTCAGGGCGAAAAAATTCTTGGGTGGAGACGAGCGGT TTAAAGTACCTTGGGTGTGGGAGGGTAGCACCGATCAGGTGTTGGTCATGGAGCGGGTAGGCGGGGTGGGTGTTG GGGATGTCAGTCATGGTAGTGCTCTCGAGGAAGGCGAGGTGGCGGTTGACGAGGAGACTGTGAGGGGTTTGAGCC AGAGGGATCGGAATGACATTGCTGCTCGAATCATTGAACTGTGCCTGAAAGAGCTCTTCGAGTTCCGGGCGATGC AGACAGATCCCAATTGGACAAACTTCCTCTGGAACCAAAAGACCAGGCAGATCGAACTCGTCGACTTTGGCGCAA CACGCACATACAGCAAAAAGTTCATGGACAGCTGGTTAAGGCTGCTCCAAGCAGCCGCCTCCGAGGACCGCCAAA CATGTATCGATGAAAGCATCAAAATCGGATACTTGACGGGCCAAGAGGACGAAGTCATGCTCGATGCACATGTTC AGTCTATGACTCTACTCGCCACTCCTTTCAAAGAAACCACTACCCAGCCGTTTGCGTTCGGACCTGGCTCGAGGT GGTCGGAAGTGACGAAGGAGATTAGGGAGTTCATTCCTGTGATGGTGAAGAGGAGGTTGACCCCCCCGCCGAAGG AGACATATAGTTTGAATAGAAAATTGAGCGGGGCGTTCCTCCTCGCTGCTAGACTCGATGCCAAGGTTGATACGA AGGAGATTTGGGATAGGATTGTTGGGAAGTACGAGTTTGGGCGTGTGGAAGAGGAGGCGGCGGAGTTGGCTAAAG ATATTGATTGATTAGCTCGACCACCGGGTTGCACTTTTCGGCACGATGGTGGAACGTTGACTAATGTATTGGGTA TACTTACTGGTGTCTTTTATAATAATCCCCCACTATTAAAGGAAAGGCT |
| Length | 2599 |
Gene
| Sequence id | CopciAB_461315.T0 |
|---|---|
| Sequence |
>CopciAB_461315.T0 GTGAATCTTTTGGTGACGACACCGACAACCGCGAAGACGGACGCGAGCTCATCGCGCTGCACGACGACAACGGGT CTCTTTTTCGTGTTCCTTATCGCGGCTATTGAGCTCTAGGACATCGGGCAACCAGGTCAACGTAAAGAAAGAAAG GCATGCCGCCAGGCCCGGCGTACAATGCTCTCGCTGTGGTCTATTCTGTCGCTCGGATCGTCGAGCATGCGGCAC GACATCGAGCTGCAGCTGGCGCTCGAACTTTGGGCGTCTCAGAGGATATACTGACGACGACACGGCCTGGACCAG CACCACAACCCGCACCAACTGCACCAACATCCTCGACCCCTTCGACTTCAACCTCGTCCGACGCTTCAGCTAGCA CAAGCCCGTCCGAGGAGAGGGTAGGAGAAGCCTCTACACAGACTCAACCAAAATCTCAGCTCTCCAAAGCAGAAC GACGACGGATACAAGCGCAACTAGAAGAGGAAGAGTTCCGAGCGATACTTGGAAAACGATGGGTTGATGGCGTAT TGGTCGACGGGTCTCTCCCATTCAGCGTTTCGCACAGCCATCCAGCTCAAATTATATCTGAAGCACCCAGGGACC CTGTGCCGCCGATACCAGAAATACCAGAACTGCCAACTCAGTCGCAGTCGCCGGCAGAGTCTTTGGAGTCGGGGA CGAAGACGTGGGTGGAGAAGGAAAGAGAGGCAGAGGAAGTTGCGAGAGTTGTGGAGGAGGTTAGCCATGAAATCG AACACGAACGCGAATTGGAAACGAAGGAAAGGCCTATACCTCAGGTGGCGCAGAAGTACCAGTCCAGTCACCAGC AGGCATCGTCCTCATCTGCAACCAGTGTTCCCACTTCAAACCCGACTGCTGCAATCGAAGCTTCTGACCCTTCAC CGACACAGGCAGCTTCTACGTCGCCTTCAGACCTTTCAGACTCTCCCTCATGCCCTCTTCCCGTCACAACCCCTA CACCACCACCCAAAAAGGTGCTTCAAGCATCTAAAGTTCCCTCGTCACGGATAGGCAGATTATTCCACTATGGAG GTACATTCAGGGCCAGCCATTCCTTGTAGTCTTTATTAACATACTCTTATACAGGTCTCGCTGCTTCACTAGGCT ATGGAGCCGCCGCTGAACTCCTTAGAGGGAGTGATAGCTCCAACGCAACAGGCAACGTCATGCTCTCAGAAGCGA ACATCAAGCGATTGGTGGGCAAGCTTACCCAGATGCGAGGGGCCGCTCTCAAGCTTGGGCAGTTCATGAGTATCC AAGGTGCTTTTCCCAGCTCATCCTCCTCGCCGCTTACTTGACGTGCTCACGCAGTTGGGTAGATACACATCTCCT CCCACCAGAGCTCGACCAGATATTCAGACGTGTCCAGGATAGCGCGCATTACATGCCAGATTGGCAAATGGAGGT TAGCCCCATGGCTGTGTGTCTGTTGCTTCCCACAATTCTAACCAGTTTATTGCGCGTAGAAAGTCCTCTCAGCGG CGTACGGTCCTAACTGGTGCAACCCTTCAAATCCCGAAGAAACTGTCTTCATGGAGTTCGAGCGGATACCGTTCG CGGCTGCGTCCATCGGTCAGGTGCACCGAGCCAGGCTAGCACCGCGACTTTGCCCACCCGAGTTGCTCGACCAGG GTAGTAAGTGGTCGAAGCGCATTCGGTTGCATTTAAGTCTCACCAACTGTTTTCTCCATAGACAAGGATGTTGGC CTAGACGTCGCAGTCAAAGTCCAATTCCCAAACATCGTAAACTCGATAAAGAGTGACTTGGGTTATGTCAAAATG CTACTCAGCGTCGGAGGTCTGCTGCCTAAAGGTCTTTTCCTGGACAGGACGATCGAGGTACGCACTCTCTTCCTC CATTTCCTTTCCCCCACTCATACACAGTGCAAATTTACAGGTGATGTCCGCGGAACTAGCAGACGAATGCTCCTA CACCCGCGAAGCCTCGTTCCTCAAGAAATTCAGGGCGAAAAAATTCTTGGGTGGAGACGAGCGGTTTAAAGTACC TTGGGTGTGGGAGGGTAGCACCGATCAGGTGTTGGTCATGGAGCGGGTAGGCGGGGTGGGTGTTGGGGATGTCAG TCATGGTAGTGCTCTCGAGGAAGGCGAGGTGGCGGTTGACGAGGAGACTGTGAGGGGTTTGAGCCAGAGGGATCG GAATGACGTGCGTATACATTCCTTGAGTGGCCTGGAGGATGCTGACAGTCGTATCTTCCCGAACAGATTGCTGCT CGAATCATTGAACTGTGCCTGAAAGAGCTCTTCGAGTTCCGGGCGATGCAGACAGATCCCAATTGGACAAACTTC CTCTGGAACCAAAAGACCAGGCAGGCGAGTACAACTCCTACACCTTTGCAACAACACGCAGAAGCCAACCGTAAT TCTCTGCTTGCAGATCGAACTCGTCGACTTTGGCGCAACACGCACATACAGCAAAAAGTTCATGGACAGCTGGTT AAGGCTGCTCCAAGCAGCCGCCTCCGAGGACCGCCAAACATGTATCGATGAAAGCATCAAAATCGGATACTTGAC GGGCCAAGAGGACGAAGTCATGCTCGATGCACATGTTCAGTCTATGACTCTACTCGCCACTCCTTTCAAAGAAAC CACTACCCAGCCGTTTGCGTTCGGACCTGGCTCGAGGTGGTCGGAAGTGACGAAGGAGATTAGGGAGTTCATTCC TGTGATGGTGAAGAGGAGGTTGACCCCCCCGCCGAAGGAGACATATAGTTTGAATAGGTACGTTTGGCGTTGGGA TCTGTTGTTTTGTTTTGTTGCACTGGTGTGGCTGATCTGGGTCGTGTTCCAGAAAATTGAGCGGGGCGTTCCTCC TCGCTGCTAGACTCGATGCCAAGGTTGATACGAAGGAGATTTGGGATAGGATTGTTGGGAAGTACGAGTTTGGGC GTGTGGAAGAGGAGGCGGCGGAGTTGGCTAAAGATATTGATTGATTAGCTCGACCACCGGGTTGCACTTTTCGGC ACGATGGTGGAACGTTGACTAATGTATTGGGTATACTTACTGGTGTCTTTTATAATAATCCCCCACTATTAAAGG AAAGGCT |
| Length | 3082 |