CopciAB_461884
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_461884 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 461884 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_2:1013280..1015650 | Strand | + |
| Gene length (nt) | 2371 | Transcript length (nt) | 1986 |
| CDS length (nt) | 1290 | Protein length (aa) | 429 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Pleurotus ostreatus PC15 | PleosPC15_2_1073610 | 41.6 | 1.652E-92 | 309 |
| Pleurotus ostreatus PC9 | PleosPC9_1_92197 | 41.6 | 1.617E-92 | 309 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21530 | 40.1 | 1.181E-90 | 304 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1381183 | 40.6 | 2.23E-90 | 303 |
| Agrocybe aegerita | Agrae_CAA7271603 | 36.9 | 4.176E-81 | 276 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_13171 | 31.6 | 1.65E-69 | 242 |
| Lentinula edodes B17 | Lened_B_1_1_3879 | 31.1 | 6.658E-68 | 237 |
| Lentinula edodes NBRC 111202 | Lenedo1_1204415 | 33 | 9.751E-65 | 228 |
| Flammulina velutipes | Flave_chr10AA00652 | 32.5 | 7.775E-41 | 157 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_153681 | 30.3 | 1.615E-37 | 147 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_461884.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 86 | 365 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000961 | AGC-kinase, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K04345 |
| K19584 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
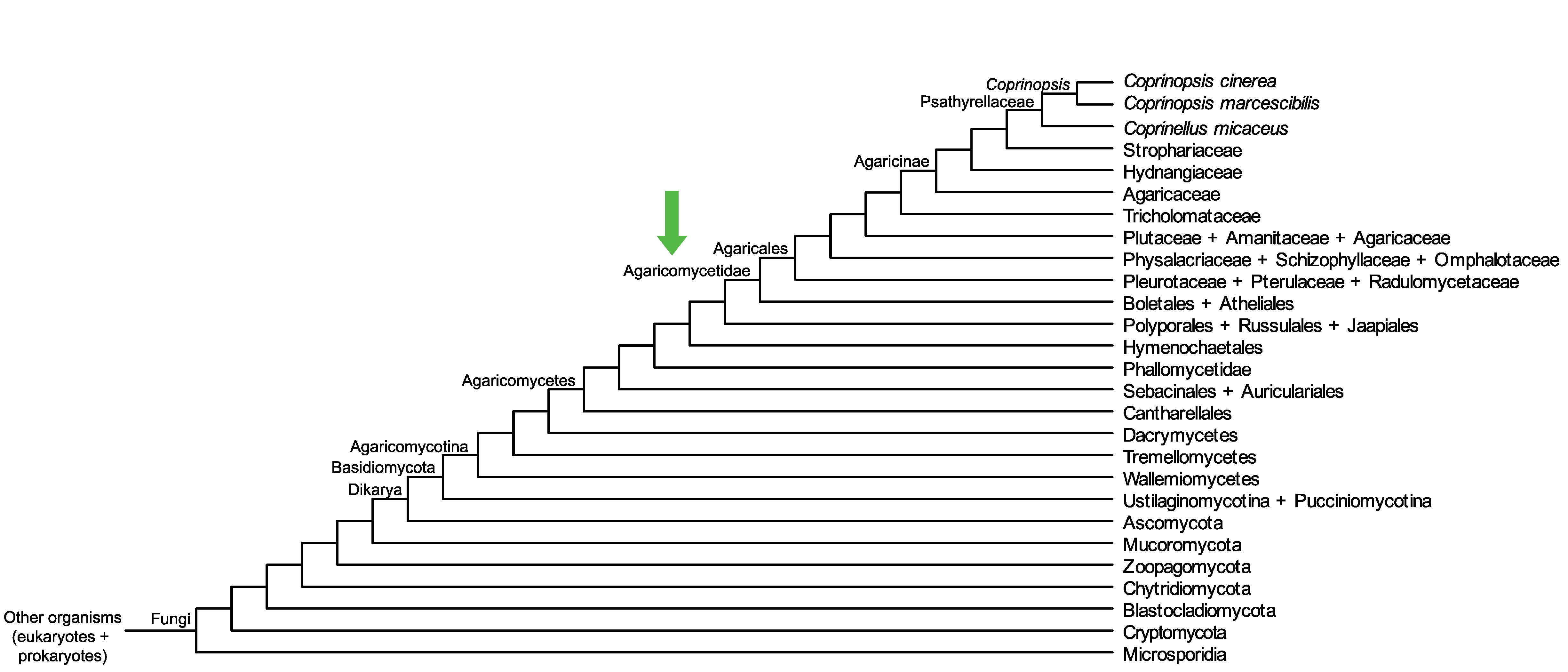
Conservation of CopciAB_461884 across fungi.
Arrow shows the origin of gene family containing CopciAB_461884.
Protein
| Sequence id | CopciAB_461884.T0 |
|---|---|
| Sequence |
>CopciAB_461884.T0 MGPVRANKSTREARNHVEAGPTQRSPPQAPYEEKKEDGYVLVAAGKLGRVYKSAYDEERFEKPVPDPELVQPVSK RPPPLGLQDLEIIKALGDGAEGRVYLVRTTRSSHPLDRPGTLYAVKTQKKKYKRQALPPEAHMLEKDNERTALAL MDWNPFIAGVIDILHDVKNIYTILELIPSGTLLSLIKSHAPMPPTIAAFYFANMFSAIEYVHALGLVHRDIKPDN FLLGEDGYLSLTDFGLTVPVDDFQVDWVDIGTPLYRPPEMDVRASPAQEKEGDEEEVDLDDRRSVDWYSSAVVLY EMVTRTVPFYGRTKLQTNALKVEKKYSFPKDIRIGSHFKDLIKSLLDPEPSNRPRPAEIQENKWLSRIDWDKLHE KQYLPPHLPKNPSASKGMWHRQPLPNPQDVPGLKVVYPPSHILHDNRFPPKLDL |
| Length | 429 |
Coding
| Sequence id | CopciAB_461884.T0 |
|---|---|
| Sequence |
>CopciAB_461884.T0 ATGGGACCCGTACGAGCCAACAAGTCGACTCGCGAGGCGAGGAATCACGTTGAGGCTGGCCCAACGCAGAGATCG CCTCCTCAGGCTCCGTACGAAGAGAAGAAAGAGGATGGTTATGTTCTGGTCGCGGCAGGAAAGCTGGGAAGAGTG TATAAGAGTGCTTATGACGAGGAGCGATTTGAGAAGCCTGTCCCAGACCCTGAACTCGTCCAACCTGTATCTAAA CGGCCACCGCCACTCGGGCTACAGGACCTTGAGATCATAAAGGCTTTGGGGGACGGAGCAGAAGGTCGGGTATAT CTCGTTCGGACAACGCGTTCAAGCCACCCTTTGGATCGGCCTGGCACCCTTTACGCTGTCAAGACGCAGAAGAAG AAATACAAACGACAAGCTCTTCCCCCCGAAGCCCATATGCTCGAGAAAGACAATGAACGTACAGCGCTAGCTCTA ATGGACTGGAACCCATTTATCGCAGGTGTTATCGACATCCTCCATGATGTCAAAAATATATACACGATTCTCGAA CTCATACCTTCTGGTACTCTCCTTTCCCTCATCAAATCACACGCCCCAATGCCACCCACCATCGCCGCGTTCTAC TTTGCCAATATGTTCTCTGCGATCGAGTACGTCCATGCATTGGGACTTGTCCACAGGGACATTAAACCAGACAAC TTCCTCCTTGGAGAGGACGGGTACTTGTCGTTGACGGACTTTGGGTTGACCGTGCCGGTTGATGATTTCCAGGTC GATTGGGTGGACATCGGAACCCCGTTGTATCGTCCACCGGAGATGGATGTTCGTGCTTCTCCTGCTCAAGAGAAG GAAGGTGACGAGGAAGAAGTAGACCTCGACGATAGACGTTCGGTTGATTGGTATTCCTCCGCCGTCGTACTTTAT GAGATGGTTACGAGGACAGTGCCGTTCTATGGCCGCACCAAACTCCAGACTAATGCTTTAAAGGTGGAGAAAAAA TATTCCTTCCCCAAGGACATCAGGATTGGAAGCCACTTCAAGGACCTCATCAAGTCTTTGCTCGACCCCGAACCC TCCAACCGGCCACGCCCGGCGGAAATCCAAGAAAACAAATGGCTAAGCCGAATCGACTGGGATAAGTTGCACGAG AAGCAATATCTGCCGCCCCATCTTCCAAAGAACCCGTCCGCCTCAAAGGGTATGTGGCATCGACAGCCCCTGCCG AATCCTCAAGACGTTCCTGGACTCAAAGTCGTCTACCCGCCTTCACACATCCTGCATGACAATCGCTTCCCGCCG AAACTAGATCTCTAG |
| Length | 1290 |
Transcript
| Sequence id | CopciAB_461884.T0 |
|---|---|
| Sequence |
>CopciAB_461884.T0 ATCTTCGACCACCATCTCATGGGACCCGTACGAGCCAACAAGTCGACTCGCGAGGCGAGGAATCACGTTGAGGCT GGCCCAACGCAGAGATCGCCTCCTCAGGCTCCGTACGAAGAGAAGAAAGAGGATGGTTATGTTCTGGTCGCGGCA GGAAAGCTGGGAAGAGTGTATAAGAGTGCTTATGACGAGGAGCGATTTGAGAAGCCTGTCCCAGACCCTGAACTC GTCCAACCTGTATCTAAACGGCCACCGCCACTCGGGCTACAGGACCTTGAGATCATAAAGGCTTTGGGGGACGGA GCAGAAGGTCGGGTATATCTCGTTCGGACAACGCGTTCAAGCCACCCTTTGGATCGGCCTGGCACCCTTTACGCT GTCAAGACGCAGAAGAAGAAATACAAACGACAAGCTCTTCCCCCCGAAGCCCATATGCTCGAGAAAGACAATGAA CGTACAGCGCTAGCTCTAATGGACTGGAACCCATTTATCGCAGGTGTTATCGACATCCTCCATGATGTCAAAAAT ATATACACGATTCTCGAACTCATACCTTCTGGTACTCTCCTTTCCCTCATCAAATCACACGCCCCAATGCCACCC ACCATCGCCGCGTTCTACTTTGCCAATATGTTCTCTGCGATCGAGTACGTCCATGCATTGGGACTTGTCCACAGG GACATTAAACCAGACAACTTCCTCCTTGGAGAGGACGGGTACTTGTCGTTGACGGACTTTGGGTTGACCGTGCCG GTTGATGATTTCCAGGTCGATTGGGTGGACATCGGAACCCCGTTGTATCGTCCACCGGAGATGGATGTTCGTGCT TCTCCTGCTCAAGAGAAGGAAGGTGACGAGGAAGAAGTAGACCTCGACGATAGACGTTCGGTTGATTGGTATTCC TCCGCCGTCGTACTTTATGAGATGGTTACGAGGACAGTGCCGTTCTATGGCCGCACCAAACTCCAGACTAATGCT TTAAAGGTGGAGAAAAAATATTCCTTCCCCAAGGACATCAGGATTGGAAGCCACTTCAAGGACCTCATCAAGTCT TTGCTCGACCCCGAACCCTCCAACCGGCCACGCCCGGCGGAAATCCAAGAAAACAAATGGCTAAGCCGAATCGAC TGGGATAAGTTGCACGAGAAGCAATATCTGCCGCCCCATCTTCCAAAGAACCCGTCCGCCTCAAAGGGTATGTGG CATCGACAGCCCCTGCCGAATCCTCAAGACGTTCCTGGACTCAAAGTCGTCTACCCGCCTTCACACATCCTGCAT GACAATCGCTTCCCGCCGAAACTAGATCTCTAGAGGTGAGAATTTGAGTTTTCCAGAGATGTAGAAGTCGCCAGT GGACGGCGAGTGGATGAAAAACGCGAATTCCGGACTGTTGCTGCGCTAGTACGGACAGTCCTCATATGACTTCAG GCCCGCACTGCGAAGACATCTGAGACCTTAGTGCTTAGTCTCCATTTGACGTGCCGAACTCTCCCATCCGAGTCT TACAGATGGAGAAACTCCCCCCGGCTCTCGAGATGTCGAACCCTGGTTGGTTCCCACTTCCTCCTTGACGGAGGA ATGAGCATCCGATGCTTTACTGTCCGGAGAGTCTGAGTACTTATGCCTATGTCGTAGAACAGGAGGACGCGTATC GGGTGCTAAGCGAACGACATGGGGCATGGTCTTTCTTATCTACGCTTCTCAACTGCTTTCTGGGCCGATTAGACG TTCTTCCTACTTACGGCTATGGTCGATCCCCTGGGTTTCTGAAATACCCACTCTATCCAGCCCCTGGTAATCCCA GGCAGTCTGTAATACCTTATCACCTGGATTTTCCATCTTTGCTTTCTTTTTTCTCAACGTCGAATTCCCTTCTAT ACAATGCAACTTGTACTAATACTTTGTCGCTTCCCAAACATCCTCTATATACATATACACCTCCTATAACCAATA CAACCGAGTACCAGTCAGATATCCTCACAGAACCTC |
| Length | 1986 |
Gene
| Sequence id | CopciAB_461884.T0 |
|---|---|
| Sequence |
>CopciAB_461884.T0 ATCTTCGACCACCATCTCATGGGACCCGTACGAGCCAACAAGTCGACTCGCGAGGCGAGGAATCACGTTGAGGCT GGCCCAACGCAGAGATCGCCTCCTCAGGCTCCGTACGAAGAGAAGAAAGAGGATGGTTATGTTCTGGTCGCGGCA GGAAAGCTGGGAAGAGTGTATAAGAGTGCTTATGACGAGGAGCGATTTGAGAAGGTGTGTGTCCTTTCGGTTGCA CCAAGTGAACGTGTTTGATTGCAGGACAGCCTGTCCCAGACCCTGAACTCGTCCAACCTGTATCTAAACGGCCAC CGCCACTCGGGCTACAGGACCTTGAGATCATAAAGGCTTTGGGTAACGCAAACTCCTGAATTTCGGAATGTGCCA CAGGTTCCTGACATGAAATCTAGGGGACGGAGCAGAAGGTCGGGTATATCTCGTTCGGACAACGCGTTCAAGCCA CCCTTTGGATCGGCCTGGCACCCTTTACGCTGTCAAGACGCAGAAGAAGAAATACAAACGACAAGCTCTTCCCGT TCGTATTTTAATTTCTGCGCTTTAGGCATGATGTCTGACCTCTCACCGAACAGCCCGAAGCCCATATGCTCGAGA AAGACAATGAACGTACAGCGCTAGCTCTAATGGACTGGAACCCATTTATCGCAGGTGTTATCGACATCCTCCATG ATGTCAAAAATATATACACGATTCTCGAACTCATACCTTCTGGTACTCTCCTTTCCCTCATCAAATCACACGCCC CAATGCCACCCACCATCGCCGCGTTCTACTTTGCCAATATGTTCTCTGCGATCGAGTACGTCCATGCATTGGGAC TTGTCCACAGGGACATTAAACCAGACAACTTCCTCCTTGGAGAGGACGGGTACTTGTCGTTGACGGACTTTGGGT TGACCGTGCCGGTTGATGATTTCCAGGTCGATTGGGTGGACATCGGAACCCCGTTGTATCGTCCACCGGAGATGG ATGTTCGTGCTTCTCCTGCTCAAGAGAAGGAAGGTGACGAGGAAGAAGTAGACCTCGACGATAGACGTTCGGTTG ATTGGTATTCCTCCGCCGTCGTACTTTATGAGATGGTTACGAGGACAGTGGTAAGTCGCCCTTTCCCATCCGCCC ATTGGGTTCCAAGCTGACCGGGCCCACCAACAGCCGTTCTATGGCCGCACCAAACTCCAGACTAATGCTTTAAAG GTGGAGAAAAAATATTCCTTCCCCAAGGACATCAGGATTGGAAGCCACTTCAAGGACCTCATCAAGTCTTTGCTC GACCCCGAACCCTCCAACCGGCCACGCCCGGCGGAAATCCAAGAAAACAAATGGCTAAGCCGAATCGACTGGGAT AAGTTGCACGAGAAGCAATATCTGGTACGTCCCCCTTTCTCCTCTCCTCTCTCCTTCTTCCCGATGGAATATTGC CATCCATATTGAATTCATCATGCGGACGATGGACTGACGTAAATCGCGGGACTATGCAGCCGCCCCATCTTCCAA AGAACCCGTCCGCCTCAAAGGGTATGTGGCATCGACAGCCCCTGCCGAATCCTCAAGACGTTCCTGGACTCAAAG TCGTCTACCCGCCTTCACACATCCTGCATGACAATCGCTTCCCGCCGAAACTAGATCTCTAGAGGTGAGAATTTG AGTTTTCCAGAGATGTAGAAGTCGCCAGTGGACGGCGAGTGGATGGTGAGACCCTTTTCCCCGCGCTTTGCTGGA GATATATATACTCATCGTGAGCCTAGAAAAACGCGAATTCCGGACTGTTGCTGCGCTAGTACGGACAGTCCTCAT ATGACTTCAGGCCCGCACTGCGAAGACATCTGAGACCTTAGTGCTTAGTCTCCATTTGACGTGCCGAACTCTCCC ATCCGAGTCTTACAGATGGAGAAACTCCCCCCGGCTCTCGAGATGTCGAACCCTGGTTGGTTCCCACTTCCTCCT TGACGGAGGAATGAGCATCCGATGCTTTACTGTCCGGAGAGTCTGAGTACTTATGCCTATGTCGTAGAACAGGAG GACGCGTATCGGGTGCTAAGCGAACGACATGGGGCATGGTCTTTCTTATCTACGCTTCTCAACTGCTTTCTGGGC CGATTAGACGTTCTTCCTACTTACGGCTATGGTCGATCCCCTGGGTTTCTGAAATACCCACTCTATCCAGCCCCT GGTAATCCCAGGCAGTCTGTAATACCTTATCACCTGGATTTTCCATCTTTGCTTTCTTTTTTCTCAACGTCGAAT TCCCTTCTATACAATGCAACTTGTACTAATACTTTGTCGCTTCCCAAACATCCTCTATATACATATACACCTCCT ATAACCAATACAACCGAGTACCAGTCAGATATCCTCACAGAACCTC |
| Length | 2371 |