CopciAB_462137
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_462137 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | SKS1 | Synonyms | 462137 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_8:547808..550916 | Strand | + |
| Gene length (nt) | 3109 | Transcript length (nt) | 3055 |
| CDS length (nt) | 2397 | Protein length (aa) | 798 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB30660 | 62.4 | 3.752E-254 | 797 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_62752 | 49.4 | 3.717E-228 | 721 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_120057 | 49.3 | 8.491E-227 | 717 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_15968 | 52 | 5.124E-194 | 622 |
| Schizophyllum commune H4-8 | Schco3_2641308 | 51 | 2.208E-185 | 597 |
| Flammulina velutipes | Flave_chr10AA01288 | 57.3 | 9.302E-184 | 592 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1482884 | 48.9 | 2.946E-166 | 541 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_157198 | 43.8 | 6.638E-164 | 534 |
| Grifola frondosa | Grifr_OBZ67312 | 44.9 | 3.806E-160 | 523 |
| Lentinula edodes NBRC 111202 | Lenedo1_1038254 | 79.7 | 3.83E-147 | 485 |
| Agrocybe aegerita | Agrae_CAA7264385 | 47 | 2.021E-145 | 480 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1061950 | 65.7 | 4.414E-141 | 467 |
| Pleurotus ostreatus PC9 | PleosPC9_1_61046 | 72.2 | 6.717E-105 | 360 |
| Lentinula edodes B17 | Lened_B_1_1_981 | 81.2 | 3.096E-101 | 349 |
| Auricularia subglabra | Aurde3_1_1101877 | 42.7 | 1.355E-65 | 242 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | SKS1 |
|---|---|
| Protein id | CopciAB_462137.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 27 | 285 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
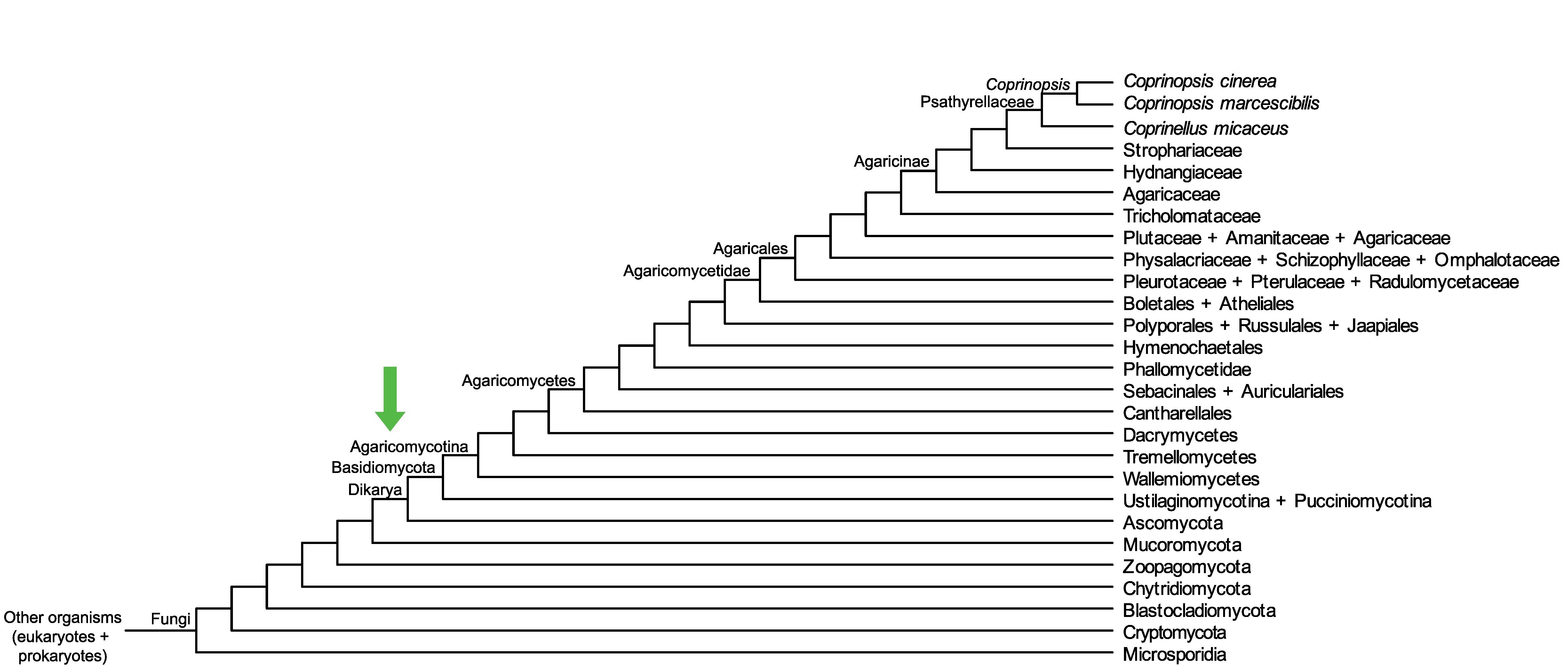
Conservation of CopciAB_462137 across fungi.
Arrow shows the origin of gene family containing CopciAB_462137.
Protein
| Sequence id | CopciAB_462137.T0 |
|---|---|
| Sequence |
>CopciAB_462137.T0 MSSKMPQPSSAVHPPVGTLIDGDSLELVEVLGVGGYGVVYRAVDARYPTGPKSYAVKCLVTSGHQSARQRQVHIR EIALHQLASAHPGVVTLHRVVEQYNHTYIIMEYASDHDLFTQILHNCRYLGNDALIKDIFLQMLDAVEYCHSLGI YHRDLKPENILCFDDGLRVAITDFGLATTDKISDEFRTGSVYHMSPECQGGAFAPGGKYSPMFNDIWSLGIILLN LATGRNPWKSATPDDPTFQAYLRDPYTFLPTVLPISAELNEILVQMLDVDWRRRMKLHEIRYAIEELTTFYSDGV IFEGSMARCPWEAGMDIDSESSANTQEVSSSPPVSPVQEQRLTSQWSKDSTSDIVFAPRSPTDDSTYGVPWPAEY SSCGATWAYESPVSSFSSEDDDHFRMDTFDRPDTPSEASTQSPPPSLPATPNDLDVTFAAQQDAKPRARLTLDTN VHKPRIVQANTSMEPSFSTDSDMMHTAIEYDPYSSLFFLATPLSSKLMGPDSAVTAVAEDREMTSPSAWRTSVVM SSPSLYSSSASSSSDDIVFSRSGTPSPEPWPLSTTYSAQVQSTPSQQCQPISPLSGMMTDSPSRPSLSPFVAHKK LRVPPTSGLSKPSSPIRVFSPPSPTTPTVYNAASGSSNDSGSSQASQLHYRFSKLFPRTNAPTPAPAAPSSTVRP ASRGRAFHAKPYPTFGRRKAGVAKAGKGDRDGKRAQQQRSVGSADGGEQGSLVASVESSSTTASACSGHDSAGTC GSASSSVSGGSCGVVVENVERKRVLGKRIATKHWFPGRFFNSGVHVGV |
| Length | 798 |
Coding
| Sequence id | CopciAB_462137.T0 |
|---|---|
| Sequence |
>CopciAB_462137.T0 ATGTCTTCGAAGATGCCTCAACCCTCCTCCGCCGTTCACCCGCCCGTTGGGACTCTCATTGACGGAGACTCCCTA GAACTCGTAGAAGTTCTCGGCGTCGGCGGCTACGGCGTCGTCTACCGTGCAGTCGACGCCCGCTATCCCACGGGT CCGAAGTCCTACGCCGTGAAGTGCCTGGTCACCTCTGGCCACCAGTCAGCGCGCCAGCGCCAAGTCCACATCCGT GAAATCGCCCTGCACCAGCTCGCCTCTGCTCACCCTGGTGTCGTCACTCTCCACCGAGTAGTGGAGCAGTACAAC CACACTTATATTATTATGGAATACGCCTCCGACCACGACCTATTTACGCAGATTCTTCACAACTGTCGATATCTC GGTAACGACGCCCTTATCAAAGACATCTTCTTGCAGATGCTCGATGCAGTGGAATATTGCCACTCGCTTGGTATT TATCATCGGGATCTCAAACCCGAGAACATCCTTTGTTTCGACGATGGCCTCCGTGTCGCCATCACCGACTTTGGT CTGGCTACCACCGACAAAATTAGCGACGAGTTTCGCACAGGAAGCGTCTATCATATGAGCCCAGAGTGCCAGGGT GGAGCTTTTGCACCTGGAGGCAAATACTCTCCCATGTTTAACGACATCTGGTCCCTTGGCATCATCCTTCTCAAT CTCGCGACTGGTCGCAACCCATGGAAGTCTGCGACTCCCGACGACCCCACTTTCCAAGCCTACCTCCGCGACCCA TATACCTTCCTTCCCACCGTCCTCCCCATCTCCGCCGAGCTCAACGAAATCCTCGTCCAGATGCTCGATGTCGAC TGGCGCCGGCGTATGAAGCTCCACGAGATCCGCTACGCCATCGAAGAGCTCACTACCTTCTACTCGGACGGTGTT ATCTTCGAAGGAAGCATGGCTCGATGCCCATGGGAAGCCGGTATGGATATCGACAGCGAGTCGTCCGCCAACACC CAGGAGGTGTCCTCCTCCCCTCCCGTCTCCCCTGTTCAGGAACAGCGTCTCACTTCGCAATGGAGCAAGGACTCC ACCTCCGACATCGTCTTTGCACCCCGCTCCCCCACCGACGACTCTACCTATGGTGTTCCATGGCCCGCCGAGTAC TCTTCTTGTGGTGCTACCTGGGCTTACGAATCCCCTGTCTCCTCGTTCTCGAGTGAAGACGATGATCACTTTCGG ATGGACACCTTTGACCGACCCGACACCCCGTCTGAAGCGTCAACGCAATCACCGCCACCTTCGCTCCCTGCTACT CCCAACGACCTTGACGTGACGTTTGCTGCCCAGCAAGACGCCAAGCCTCGTGCTCGCCTCACTCTCGACACCAAT GTCCACAAGCCTCGCATCGTCCAGGCCAATACGAGCATGGAGCCTTCGTTCTCTACCGACAGCGACATGATGCAT ACCGCGATCGAGTATGACCCGTACTCGTCGCTCTTCTTCCTCGCCACTCCACTGTCGTCCAAGCTCATGGGTCCC GACTCAGCCGTCACGGCTGTTGCAGAGGACAGGGAGATGACCTCTCCCAGCGCTTGGAGGACGTCGGTCGTCATG TCGTCTCCCTCCCTCTACTCTTCTTCGGCAAGTTCTTCCTCCGACGACATTGTGTTCTCTCGATCTGGCACGCCC TCTCCTGAACCTTGGCCTCTGTCAACAACGTATTCCGCTCAGGTACAGTCCACGCCTTCTCAGCAATGTCAGCCC ATTTCTCCGCTCTCTGGCATGATGACCGACTCCCCTTCAAGGCCCTCTCTCTCGCCCTTCGTCGCCCACAAGAAG CTCAGGGTCCCACCAACCTCCGGCCTCTCCAAGCCCAGCTCCCCCATCCGAGTCTTCTCTCCGCCCTCACCTACA ACTCCAACTGTCTACAACGCCGCCTCCGGCAGCAGCAACGACAGCGGAAGCAGCCAAGCCTCCCAACTCCACTAT CGATTCTCCAAGCTCTTCCCTCGCACGAACGCACCGACGCCTGCACCGGCTGCACCAAGTTCGACAGTCCGTCCT GCTTCACGTGGGCGCGCTTTCCACGCTAAACCTTATCCAACCTTTGGTCGCCGTAAAGCGGGCGTGGCAAAAGCT GGGAAGGGTGACCGTGACGGGAAGAGGGCGCAGCAACAGCGATCCGTTGGGAGCGCAGATGGCGGGGAGCAAGGT TCCTTGGTGGCGTCCGTCGAGTCTTCTTCAACGACGGCCAGTGCCTGCTCGGGCCATGACAGCGCCGGAACTTGT GGGTCTGCGAGCTCCTCAGTCTCCGGTGGGAGTTGTGGGGTTGTAGTCGAGAACGTGGAGAGGAAGAGAGTGCTT GGGAAGAGGATTGCGACGAAGCACTGGTTTCCGGGCAGGTTCTTCAACTCGGGGGTGCATGTTGGCGTCTGA |
| Length | 2397 |
Transcript
| Sequence id | CopciAB_462137.T0 |
|---|---|
| Sequence |
>CopciAB_462137.T0 ACCCTTCGAAGTTGCACTGGTGCTAACTCCCCTCCTCCCTCCCTCTTCCTCCCTCGGACCCTTTTCCCTCTAGTT ACGCCCCTATCACGAGTCCATGTCTTCGAAGATGCCTCAACCCTCCTCCGCCGTTCACCCGCCCGTTGGGACTCT CATTGACGGAGACTCCCTAGAACTCGTAGAAGTTCTCGGCGTCGGCGGCTACGGCGTCGTCTACCGTGCAGTCGA CGCCCGCTATCCCACGGGTCCGAAGTCCTACGCCGTGAAGTGCCTGGTCACCTCTGGCCACCAGTCAGCGCGCCA GCGCCAAGTCCACATCCGTGAAATCGCCCTGCACCAGCTCGCCTCTGCTCACCCTGGTGTCGTCACTCTCCACCG AGTAGTGGAGCAGTACAACCACACTTATATTATTATGGAATACGCCTCCGACCACGACCTATTTACGCAGATTCT TCACAACTGTCGATATCTCGGTAACGACGCCCTTATCAAAGACATCTTCTTGCAGATGCTCGATGCAGTGGAATA TTGCCACTCGCTTGGTATTTATCATCGGGATCTCAAACCCGAGAACATCCTTTGTTTCGACGATGGCCTCCGTGT CGCCATCACCGACTTTGGTCTGGCTACCACCGACAAAATTAGCGACGAGTTTCGCACAGGAAGCGTCTATCATAT GAGCCCAGAGTGCCAGGGTGGAGCTTTTGCACCTGGAGGCAAATACTCTCCCATGTTTAACGACATCTGGTCCCT TGGCATCATCCTTCTCAATCTCGCGACTGGTCGCAACCCATGGAAGTCTGCGACTCCCGACGACCCCACTTTCCA AGCCTACCTCCGCGACCCATATACCTTCCTTCCCACCGTCCTCCCCATCTCCGCCGAGCTCAACGAAATCCTCGT CCAGATGCTCGATGTCGACTGGCGCCGGCGTATGAAGCTCCACGAGATCCGCTACGCCATCGAAGAGCTCACTAC CTTCTACTCGGACGGTGTTATCTTCGAAGGAAGCATGGCTCGATGCCCATGGGAAGCCGGTATGGATATCGACAG CGAGTCGTCCGCCAACACCCAGGAGGTGTCCTCCTCCCCTCCCGTCTCCCCTGTTCAGGAACAGCGTCTCACTTC GCAATGGAGCAAGGACTCCACCTCCGACATCGTCTTTGCACCCCGCTCCCCCACCGACGACTCTACCTATGGTGT TCCATGGCCCGCCGAGTACTCTTCTTGTGGTGCTACCTGGGCTTACGAATCCCCTGTCTCCTCGTTCTCGAGTGA AGACGATGATCACTTTCGGATGGACACCTTTGACCGACCCGACACCCCGTCTGAAGCGTCAACGCAATCACCGCC ACCTTCGCTCCCTGCTACTCCCAACGACCTTGACGTGACGTTTGCTGCCCAGCAAGACGCCAAGCCTCGTGCTCG CCTCACTCTCGACACCAATGTCCACAAGCCTCGCATCGTCCAGGCCAATACGAGCATGGAGCCTTCGTTCTCTAC CGACAGCGACATGATGCATACCGCGATCGAGTATGACCCGTACTCGTCGCTCTTCTTCCTCGCCACTCCACTGTC GTCCAAGCTCATGGGTCCCGACTCAGCCGTCACGGCTGTTGCAGAGGACAGGGAGATGACCTCTCCCAGCGCTTG GAGGACGTCGGTCGTCATGTCGTCTCCCTCCCTCTACTCTTCTTCGGCAAGTTCTTCCTCCGACGACATTGTGTT CTCTCGATCTGGCACGCCCTCTCCTGAACCTTGGCCTCTGTCAACAACGTATTCCGCTCAGGTACAGTCCACGCC TTCTCAGCAATGTCAGCCCATTTCTCCGCTCTCTGGCATGATGACCGACTCCCCTTCAAGGCCCTCTCTCTCGCC CTTCGTCGCCCACAAGAAGCTCAGGGTCCCACCAACCTCCGGCCTCTCCAAGCCCAGCTCCCCCATCCGAGTCTT CTCTCCGCCCTCACCTACAACTCCAACTGTCTACAACGCCGCCTCCGGCAGCAGCAACGACAGCGGAAGCAGCCA AGCCTCCCAACTCCACTATCGATTCTCCAAGCTCTTCCCTCGCACGAACGCACCGACGCCTGCACCGGCTGCACC AAGTTCGACAGTCCGTCCTGCTTCACGTGGGCGCGCTTTCCACGCTAAACCTTATCCAACCTTTGGTCGCCGTAA AGCGGGCGTGGCAAAAGCTGGGAAGGGTGACCGTGACGGGAAGAGGGCGCAGCAACAGCGATCCGTTGGGAGCGC AGATGGCGGGGAGCAAGGTTCCTTGGTGGCGTCCGTCGAGTCTTCTTCAACGACGGCCAGTGCCTGCTCGGGCCA TGACAGCGCCGGAACTTGTGGGTCTGCGAGCTCCTCAGTCTCCGGTGGGAGTTGTGGGGTTGTAGTCGAGAACGT GGAGAGGAAGAGAGTGCTTGGGAAGAGGATTGCGACGAAGCACTGGTTTCCGGGCAGGTTCTTCAACTCGGGGGT GCATGTTGGCGTCTGACGGACTCTTTCCCTCTCTTGCTTCGACTCTTTACCCTTCGTTCTTCGCATTCCATTGGG CACGCTTTTCTTTCAATTTCACTGGCTTTTTTTCCATCCTTCTTCCATTTTATCCATGCGTCGAAACAACGCCTC TTTTGCATCATCATCATACATCCGCATCATCATCGACATCCAGCATCAGCATTTCATCGTTAGAAGAAAAACTCA TTTGAACGACGCTTTACGAGAAAAAAACACATACGAAACTTTATGACTTTTATGAAAGCATTAGGCATTTCATCG TCATCACCATCATCACACCGCATTGCATATGTATTTTTATTAGCATGGATTTTCTTTCTCTCTTCTTCATCTGCA TCTCTCCGCTCCTTTATCTCCTTTCTTCTTGTTTCTTATCTGTTGTACTCTCGGATGCGTTAGGCATCGACGTTG TTGTACATTTCGGCAACCCTAGTGACCGAGCATTGTGTAGTGTAAAAACCGGAATATGTCCCTTGTTGTATGTGT AGTTCAAAGTGGATTGTACACAATGTAATCGAATCGAAATTGTACTTCAATAACC |
| Length | 3055 |
Gene
| Sequence id | CopciAB_462137.T0 |
|---|---|
| Sequence |
>CopciAB_462137.T0 ACCCTTCGAAGTTGCACTGGTGCTAACTCCCCTCCTCCCTCCCTCTTCCTCCCTCGGACCCTTTTCCCTCTAGTT ACGCCCCTATCACGAGTCCATGTCTTCGAAGATGCCTCAACCCTCCTCCGCCGTTCACCCGCCCGTTGGGACTCT CATTGACGGAGACTCCCTAGAACTCGTAGAAGTTCTCGGCGTCGGCGGCTACGGCGTCGTCTACCGTGCAGTCGA CGCCCGCTATCCCACGGGTCCGAAGTCCTACGCCGTGAAGTGCCTGGTCACCTCTGGCCACCAGTCAGCGCGCCA GCGCCAAGTCCACATCCGTGAAATCGCCCTGCACCAGCTCGCCTCTGCTCACCCTGGTGTCGTCACTCTCCACCG AGTAGTGGAGCAGTACAACCACACTTATATTATTATGGAATACGCCTCCGACCACGACCTATTTACGCAGATTCT TCACAACTGTCGATATCTCGGTAACGACGCCCTTATCAAAGACATCTTCTTGCAGATGCTCGATGCAGTGGAATA TTGCCACTCGCTTGGTATTTATCATCGGGATCTCAAACCCGAGAACATCCTTTGTTTCGACGATGGCCTCCGTGT CGCCATCACCGACTTTGGTCTGGCTACCACCGACAAAATTAGCGACGAGTTTCGCACAGGAAGCGTCTATCATAT GAGCCCAGGTATGTTCTTACCTCGTCGTTTTTCCCTTACGTTTTCCTTACATTCGATTTCAGAGTGCCAGGGTGG AGCTTTTGCACCTGGAGGCAAATACTCTCCCATGTTTAACGACATCTGGTCCCTTGGCATCATCCTTCTCAATCT CGCGACTGGTCGCAACCCATGGAAGTCTGCGACTCCCGACGACCCCACTTTCCAAGCCTACCTCCGCGACCCATA TACCTTCCTTCCCACCGTCCTCCCCATCTCCGCCGAGCTCAACGAAATCCTCGTCCAGATGCTCGATGTCGACTG GCGCCGGCGTATGAAGCTCCACGAGATCCGCTACGCCATCGAAGAGCTCACTACCTTCTACTCGGACGGTGTTAT CTTCGAAGGAAGCATGGCTCGATGCCCATGGGAAGCCGGTATGGATATCGACAGCGAGTCGTCCGCCAACACCCA GGAGGTGTCCTCCTCCCCTCCCGTCTCCCCTGTTCAGGAACAGCGTCTCACTTCGCAATGGAGCAAGGACTCCAC CTCCGACATCGTCTTTGCACCCCGCTCCCCCACCGACGACTCTACCTATGGTGTTCCATGGCCCGCCGAGTACTC TTCTTGTGGTGCTACCTGGGCTTACGAATCCCCTGTCTCCTCGTTCTCGAGTGAAGACGATGATCACTTTCGGAT GGACACCTTTGACCGACCCGACACCCCGTCTGAAGCGTCAACGCAATCACCGCCACCTTCGCTCCCTGCTACTCC CAACGACCTTGACGTGACGTTTGCTGCCCAGCAAGACGCCAAGCCTCGTGCTCGCCTCACTCTCGACACCAATGT CCACAAGCCTCGCATCGTCCAGGCCAATACGAGCATGGAGCCTTCGTTCTCTACCGACAGCGACATGATGCATAC CGCGATCGAGTATGACCCGTACTCGTCGCTCTTCTTCCTCGCCACTCCACTGTCGTCCAAGCTCATGGGTCCCGA CTCAGCCGTCACGGCTGTTGCAGAGGACAGGGAGATGACCTCTCCCAGCGCTTGGAGGACGTCGGTCGTCATGTC GTCTCCCTCCCTCTACTCTTCTTCGGCAAGTTCTTCCTCCGACGACATTGTGTTCTCTCGATCTGGCACGCCCTC TCCTGAACCTTGGCCTCTGTCAACAACGTATTCCGCTCAGGTACAGTCCACGCCTTCTCAGCAATGTCAGCCCAT TTCTCCGCTCTCTGGCATGATGACCGACTCCCCTTCAAGGCCCTCTCTCTCGCCCTTCGTCGCCCACAAGAAGCT CAGGGTCCCACCAACCTCCGGCCTCTCCAAGCCCAGCTCCCCCATCCGAGTCTTCTCTCCGCCCTCACCTACAAC TCCAACTGTCTACAACGCCGCCTCCGGCAGCAGCAACGACAGCGGAAGCAGCCAAGCCTCCCAACTCCACTATCG ATTCTCCAAGCTCTTCCCTCGCACGAACGCACCGACGCCTGCACCGGCTGCACCAAGTTCGACAGTCCGTCCTGC TTCACGTGGGCGCGCTTTCCACGCTAAACCTTATCCAACCTTTGGTCGCCGTAAAGCGGGCGTGGCAAAAGCTGG GAAGGGTGACCGTGACGGGAAGAGGGCGCAGCAACAGCGATCCGTTGGGAGCGCAGATGGCGGGGAGCAAGGTTC CTTGGTGGCGTCCGTCGAGTCTTCTTCAACGACGGCCAGTGCCTGCTCGGGCCATGACAGCGCCGGAACTTGTGG GTCTGCGAGCTCCTCAGTCTCCGGTGGGAGTTGTGGGGTTGTAGTCGAGAACGTGGAGAGGAAGAGAGTGCTTGG GAAGAGGATTGCGACGAAGCACTGGTTTCCGGGCAGGTTCTTCAACTCGGGGGTGCATGTTGGCGTCTGACGGAC TCTTTCCCTCTCTTGCTTCGACTCTTTACCCTTCGTTCTTCGCATTCCATTGGGCACGCTTTTCTTTCAATTTCA CTGGCTTTTTTTCCATCCTTCTTCCATTTTATCCATGCGTCGAAACAACGCCTCTTTTGCATCATCATCATACAT CCGCATCATCATCGACATCCAGCATCAGCATTTCATCGTTAGAAGAAAAACTCATTTGAACGACGCTTTACGAGA AAAAAACACATACGAAACTTTATGACTTTTATGAAAGCATTAGGCATTTCATCGTCATCACCATCATCACACCGC ATTGCATATGTATTTTTATTAGCATGGATTTTCTTTCTCTCTTCTTCATCTGCATCTCTCCGCTCCTTTATCTCC TTTCTTCTTGTTTCTTATCTGTTGTACTCTCGGATGCGTTAGGCATCGACGTTGTTGTACATTTCGGCAACCCTA GTGACCGAGCATTGTGTAGTGTAAAAACCGGAATATGTCCCTTGTTGTATGTGTAGTTCAAAGTGGATTGTACAC AATGTAATCGAATCGAAATTGTACTTCAATAACC |
| Length | 3109 |