CopciAB_469209
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_469209 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 469209 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_13:915874..918504 | Strand | + |
| Gene length (nt) | 2631 | Transcript length (nt) | 2319 |
| CDS length (nt) | 1269 | Protein length (aa) | 422 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7263799 | 59.2 | 4.405E-149 | 473 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB22353 | 56.7 | 3.417E-144 | 459 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_116779 | 57.4 | 4.962E-144 | 458 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_196011 | 57.2 | 2.338E-143 | 456 |
| Flammulina velutipes | Flave_chr09AA00209 | 57.5 | 1.067E-136 | 437 |
| Schizophyllum commune H4-8 | Schco3_2629707 | 56.1 | 3.158E-135 | 433 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_56516 | 55.4 | 1.478E-132 | 425 |
| Pleurotus ostreatus PC15 | PleosPC15_2_42079 | 50.3 | 1.652E-120 | 390 |
| Pleurotus ostreatus PC9 | PleosPC9_1_100485 | 50.3 | 1.617E-120 | 390 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1506698 | 42.9 | 1.03E-118 | 385 |
| Lentinula edodes B17 | Lened_B_1_1_671 | 45.3 | 8.633E-108 | 353 |
| Lentinula edodes NBRC 111202 | Lenedo1_149370 | 45.3 | 1.091E-107 | 353 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_12742 | 45.3 | 1.083E-107 | 353 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_469209.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14016 | STKc_CK1 | - | 9 | 360 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 10 | 143 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
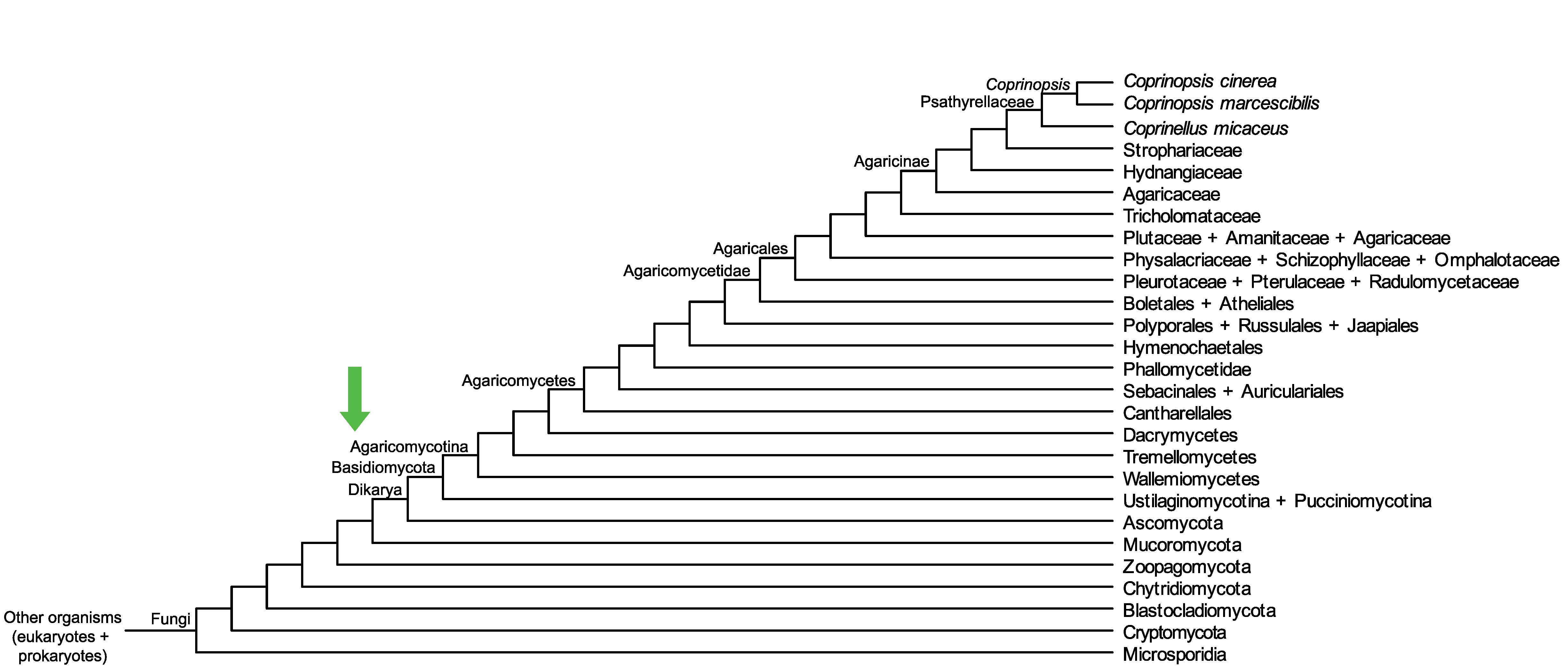
Conservation of CopciAB_469209 across fungi.
Arrow shows the origin of gene family containing CopciAB_469209.
Protein
| Sequence id | CopciAB_469209.T0 |
|---|---|
| Sequence |
>CopciAB_469209.T0 MVLTTYAGKYRFEEEIANGGCGTVFLGVHTTAGKQVAIKLEPTHPDHHPSHPSPLKTESKIYKSLSGGEGVPWII WSGRQGDYNVMVTDLLGPSLEDLFKMCNRHFSMKTVLLLADQLISRIEFVHSHSIVHRDIKPANFVMSVAPQTPC SCSTSHSNANSTAPSPTSPSSSLPDPVEGDVVKPVRSRLPSPPASSRPCPHHPLVHIIDFGLAKKFRDSVSQIHI PFSQHPTGLHGVGTSLFASINTHLGMEASRRDDLESLAYMLIYFLRGTLPWRKMRAPSNLPSSILAAISNSEERR EAQEHYNPVSATWDLIRDSKLEHEATLTEGLPEEFHVLYTYARNLEFDDLPDYEGLRNCFRGLAARLGIEYDGVF DWSVKGPGEGGSKGPGIASKKIRRGRQCLACDARAKAKAEEAERRKS |
| Length | 422 |
Coding
| Sequence id | CopciAB_469209.T0 |
|---|---|
| Sequence |
>CopciAB_469209.T0 ATGGTTCTTACTACTTATGCTGGCAAATACAGGTTTGAAGAAGAAATAGCAAATGGAGGCTGTGGAACCGTGTTT CTGGGCGTGCATACCACAGCAGGAAAACAAGTGGCAATCAAGCTAGAGCCAACCCACCCTGATCACCACCCGTCG CACCCATCGCCGCTCAAAACAGAGTCCAAAATATACAAATCCCTCTCTGGCGGTGAAGGCGTACCATGGATTATT TGGAGCGGTCGACAGGGTGACTACAACGTTATGGTCACGGATCTCCTGGGTCCCAGTCTAGAAGACCTCTTCAAG ATGTGCAATCGCCATTTTAGTATGAAGACCGTCCTCTTACTGGCAGATCAATTGATCTCCCGCATTGAATTTGTG CATTCGCACTCGATCGTCCATCGCGATATCAAGCCTGCGAACTTTGTCATGTCGGTGGCGCCACAAACACCATGC TCCTGTTCAACATCACATTCGAATGCCAACTCAACGGCACCGTCCCCAACTTCGCCCTCCTCATCCTTACCTGAC CCTGTGGAAGGTGATGTCGTCAAGCCCGTAAGATCAAGGCTGCCAAGTCCTCCGGCATCGTCGCGACCGTGTCCG CACCATCCCCTCGTCCACATCATTGACTTCGGCCTGGCCAAGAAGTTCCGAGACTCGGTATCGCAAATCCACATC CCCTTTTCTCAACACCCGACAGGCCTCCATGGGGTCGGAACCTCCCTATTCGCGTCAATAAACACTCACCTCGGC ATGGAAGCTTCGCGAAGAGATGACCTTGAATCCCTCGCCTACATGCTCATCTACTTCCTTCGTGGGACTTTGCCT TGGAGAAAGATGCGCGCCCCTTCCAACCTTCCTTCGTCGATTTTGGCGGCCATTTCGAATAGTGAAGAGCGACGT GAGGCTCAAGAGCATTATAACCCCGTCAGTGCTACGTGGGACCTGATCCGCGACAGCAAGCTCGAGCATGAAGCG ACCTTGACGGAGGGTCTTCCCGAGGAGTTCCACGTGCTGTACACCTACGCGCGTAACTTGGAGTTTGACGACCTG CCCGACTACGAGGGGTTGCGAAATTGTTTCCGTGGTCTTGCTGCACGTCTTGGTATCGAGTACGACGGTGTTTTC GATTGGAGCGTAAAGGGTCCTGGGGAAGGAGGCAGCAAGGGCCCAGGAATTGCATCAAAGAAAATCCGTCGTGGA CGACAATGCTTGGCCTGCGATGCGCGCGCGAAAGCTAAAGCCGAGGAGGCGGAGAGGAGGAAGTCGTAA |
| Length | 1269 |
Transcript
| Sequence id | CopciAB_469209.T0 |
|---|---|
| Sequence |
>CopciAB_469209.T0 ACCAACCTTTTCCCTCGTCCCCAACGTTCCTCTCAACCACCAAACGCGCGCTCGCGGTTTCTTCGCGACGTCCGC AAGATGGTTCTTACTACTTATGCTGGCAAATACAGGTTTGAAGAAGAAATAGCAAATGGAGGCTGTGGAACCGTG TTTCTGGGCGTGCATACCACAGCAGGAAAACAAGTGGCAATCAAGCTAGAGCCAACCCACCCTGATCACCACCCG TCGCACCCATCGCCGCTCAAAACAGAGTCCAAAATATACAAATCCCTCTCTGGCGGTGAAGGCGTACCATGGATT ATTTGGAGCGGTCGACAGGGTGACTACAACGTTATGGTCACGGATCTCCTGGGTCCCAGTCTAGAAGACCTCTTC AAGATGTGCAATCGCCATTTTAGTATGAAGACCGTCCTCTTACTGGCAGATCAATTGATCTCCCGCATTGAATTT GTGCATTCGCACTCGATCGTCCATCGCGATATCAAGCCTGCGAACTTTGTCATGTCGGTGGCGCCACAAACACCA TGCTCCTGTTCAACATCACATTCGAATGCCAACTCAACGGCACCGTCCCCAACTTCGCCCTCCTCATCCTTACCT GACCCTGTGGAAGGTGATGTCGTCAAGCCCGTAAGATCAAGGCTGCCAAGTCCTCCGGCATCGTCGCGACCGTGT CCGCACCATCCCCTCGTCCACATCATTGACTTCGGCCTGGCCAAGAAGTTCCGAGACTCGGTATCGCAAATCCAC ATCCCCTTTTCTCAACACCCGACAGGCCTCCATGGGGTCGGAACCTCCCTATTCGCGTCAATAAACACTCACCTC GGCATGGAAGCTTCGCGAAGAGATGACCTTGAATCCCTCGCCTACATGCTCATCTACTTCCTTCGTGGGACTTTG CCTTGGAGAAAGATGCGCGCCCCTTCCAACCTTCCTTCGTCGATTTTGGCGGCCATTTCGAATAGTGAAGAGCGA CGTGAGGCTCAAGAGCATTATAACCCCGTCAGTGCTACGTGGGACCTGATCCGCGACAGCAAGCTCGAGCATGAA GCGACCTTGACGGAGGGTCTTCCCGAGGAGTTCCACGTGCTGTACACCTACGCGCGTAACTTGGAGTTTGACGAC CTGCCCGACTACGAGGGGTTGCGAAATTGTTTCCGTGGTCTTGCTGCACGTCTTGGTATCGAGTACGACGGTGTT TTCGATTGGAGCGTAAAGGGTCCTGGGGAAGGAGGCAGCAAGGGCCCAGGAATTGCATCAAAGAAAATCCGTCGT GGACGACAATGCTTGGCCTGCGATGCGCGCGCGAAAGCTAAAGCCGAGGAGGCGGAGAGGAGGAAGTCGTAAAGA GCGGCAGGCGTGTAGGGTGTGTTTGTGGTGGGGATGGTGGGGGTTGCTTTCCTGCCGCTTTCCTGGGGGAGAATC CTGGACCCAGGAGTAGCGTGCGTTGTTTTGGGGGGTCGAACTTTTATAGGCCACCTTCTTGTCCTCGTGCATCTC GCGATGCAACCCCTATGGATGATGGGTTGCCTCTTCGCATGGCAATGCCCTCAGCCTACTGGGCTCTGACGATTC CTGCGGGATAAGCACGGATCACGCTCGCCACGCGTCGATGTTAGTGCACGAAGCAGATCGATAACCGTTAGACCA CCTGGTAGCCGCACCTTCCGCTTCGGTTTGGGAGGGCGCATATCGGCTGTATCTCTCCCTCTTCCCATTGTCAGT CCATCATATCCTCTCTTCCTCTTTGCTCTCTCGTCATGCCCCGTTTTTATTACCCGTTGCATTGCTTTCGCTCTT GACTTTTTCCTTTGTCACGGTCGAGCGCGGACGGTTCTTACCAAGTCCCATTGGCTGGAAAAGTAATTGTACTTT TATATCCTTTCTTTTGTGTTTGGGCATTACGTTCAACTTACACCATCCTCGACTCTTTTCTTCGGCACTCATTGT ACTTTAGAATTTAGTACTTTCCTGGTTTTGTCGTGGGGGTGTTTTCAGGTTGACCTGTGACATTCGCTCCATTCA TCATTTCATCTTTACCTGTCCCATTTTTTGCATCAGCATCCCTTTGCATTCGTGCATCATCACTACCCCAGTACA TATGGCATGTTCTATCATCATCCCCATTCCTCCCTTTCCTCTCGAGACTCTTTTTCTGCTTTCTGCATCATTGAA CTTTGCATTCAGTGTTGTTTTATTACCCCATCCCATTTTATCCCACTCGTCTCATCCCCAAGAGCGTAGCGCTAC ATGTAGTATTATAATTCTAGTTCCCTGTATCTGATGTGTCGTATTTATAGACGGTTTCTCTCTCCCTGT |
| Length | 2319 |
Gene
| Sequence id | CopciAB_469209.T0 |
|---|---|
| Sequence |
>CopciAB_469209.T0 ACCAACCTTTTCCCTCGTCCCCAACGTTCCTCTCAACCACCAAACGCGCGCTCGCGGTTTCTTCGCGACGTCCGC AAGATGGTTCTTACTACTTATGCTGGCAAATACAGGTTTGAAGAAGAAATAGCAAATGGAGGCTGTGGTGAGTGA ACCGGGTCCTAATCCCCACTATGTCATTGGCACACCCCTTTGCTGACCCGCTATTGGTTTCAAGGAACCGTGTTT CTGGGCGTGCATACCACAGCAGGAAAACAAGTGGCAATCAAGCTAGAGCCAACCCACCCTGATCACCACCCGTCG CACCCATCGCCGCTCAAAACAGAGTCCAAAATATACAAATCCCTCTCTGGCGGTGAAGGCGTACCATGGATTATT TGGAGCGGTCGACAGGGTGACTACAACGTTATGGTCACGGATCTCCTGGGTCCCAGTCTAGAAGACCTCTTCAAG ATGTGCAATCGCCATTTTAGTATGAAGACCGTCCTCTTACTGGCAGATCAATTGGTGAGCTTCCCAAAGCTGGAA ATACGCCACGCTGGAAGACCATTGGGCTGTCGCTATACGAGCTGGTTCCATCGTGTAGTCTCCTGCACCTCTGCA ATACCTCAGACGATGACTGGCGCTGATGGTTGCAGTGGTGGTGGTGGCATATTGCCGTGGACACACTTTTGCATA ACGAGGCACCCTCTTCCCGCTTGGTTAACTGCCGTATGAGTCCATTGCTAACTTTGAGCTTAATTTCAGATCTCC CGCATTGAATTTGTGCATTCGCACTCGATCGTCCATCGCGATATCAAGCCTGCGAACTTTGTCATGTCGGTGGCG CCACAAACACCATGCTCCTGTTCAACATCACATTCGAATGCCAACTCAACGGCACCGTCCCCAACTTCGCCCTCC TCATCCTTACCTGACCCTGTGGAAGGTGATGTCGTCAAGCCCGTAAGATCAAGGCTGCCAAGTCCTCCGGCATCG TCGCGACCGTGTCCGCACCATCCCCTCGTCCACATCATTGACTTCGGCCTGGCCAAGAAGTTCCGAGACTCGGTA TCGCAAATCCACATCCCCTTTTCTCAACACCCGACAGGCCTCCATGGGGTCGGAACCTCCCTATTCGCGTCAATA AACACTCACCTCGGCATGGAAGCTTCGCGAAGAGATGACCTTGAATCCCTCGCCTACATGCTCATCTACTTCCTT CGTGGGACTTTGCCTTGGAGAAAGATGCGCGCCCCTTCCAACCTTCCTTCGTCGATTTTGGCGGCCATTTCGAAT AGTGAAGAGCGACGTGAGGCTCAAGAGCATTATAACCCCGTCAGTGCTACGTGGGACCTGATCCGCGACAGCAAG CTCGAGCATGAAGCGACCTTGACGGAGGGTCTTCCCGAGGAGTTCCACGTGCTGTACACCTACGCGCGTAACTTG GAGTTTGACGACCTGCCCGACTACGAGGGGTTGCGAAATTGTTTCCGTGGTCTTGCTGCACGTCTTGGTATCGAG TACGACGGTGTTTTCGATTGGAGCGTAAAGGGTCCTGGGGAAGGAGGCAGCAAGGGCCCAGGAATTGCATCAAAG AAAATCCGTCGTGGACGACAATGCTTGGCCTGCGATGCGCGCGCGAAAGCTAAAGCCGAGGAGGCGGAGAGGAGG AAGTCGTAAAGAGCGGCAGGCGTGTAGGGTGTGTTTGTGGTGGGGATGGTGGGGGTTGCTTTCCTGCCGCTTTCC TGGGGGAGAATCCTGGACCCAGGAGTAGCGTGCGTTGTTTTGGGGGGTCGAACTTTTATAGGCCACCTTCTTGTC CTCGTGCATCTCGCGATGCAACCCCTATGGATGATGGGTTGCCTCTTCGCATGGCAATGCCCTCAGCCTACTGGG CTCTGACGATTCCTGCGGGATAAGCACGGATCACGCTCGCCACGCGTCGATGTTAGTGCACGAAGCAGATCGATA ACCGTTAGACCACCTGGTAGCCGCACCTTCCGCTTCGGTTTGGGAGGGCGCATATCGGCTGTATCTCTCCCTCTT CCCATTGTCAGTCCATCATATCCTCTCTTCCTCTTTGCTCTCTCGTCATGCCCCGTTTTTATTACCCGTTGCATT GCTTTCGCTCTTGACTTTTTCCTTTGTCACGGTCGAGCGCGGACGGTTCTTACCAAGTCCCATTGGCTGGAAAAG TAATTGTACTTTTATATCCTTTCTTTTGTGTTTGGGCATTACGTTCAACTTACACCATCCTCGACTCTTTTCTTC GGCACTCATTGTACTTTAGAATTTAGTACTTTCCTGGTTTTGTCGTGGGGGTGTTTTCAGGTTGACCTGTGACAT TCGCTCCATTCATCATTTCATCTTTACCTGTCCCATTTTTTGCATCAGCATCCCTTTGCATTCGTGCATCATCAC TACCCCAGTACATATGGCATGTTCTATCATCATCCCCATTCCTCCCTTTCCTCTCGAGACTCTTTTTCTGCTTTC TGCATCATTGAACTTTGCATTCAGTGTTGTTTTATTACCCCATCCCATTTTATCCCACTCGTCTCATCCCCAAGA GCGTAGCGCTACATGTAGTATTATAATTCTAGTTCCCTGTATCTGATGTGTCGTATTTATAGACGGTTTCTCTCT CCCTGT |
| Length | 2631 |