CopciAB_469373
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_469373 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 469373 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_4:1555624..1557397 | Strand | + |
| Gene length (nt) | 1774 | Transcript length (nt) | 1554 |
| CDS length (nt) | 1359 | Protein length (aa) | 452 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Saccharomyces cerevisiae | YGL179C_TOS3 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_204748 | 50.1 | 2.284E-139 | 446 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_71777 | 50.1 | 5.27E-139 | 445 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB25022 | 51.3 | 3.94E-138 | 443 |
| Agrocybe aegerita | Agrae_CAA7259115 | 52.3 | 4.115E-136 | 437 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1461132 | 50.3 | 3.981E-126 | 408 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1056567 | 50.5 | 1.655E-125 | 406 |
| Pleurotus ostreatus PC9 | PleosPC9_1_77424 | 50.5 | 1.62E-125 | 406 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18218 | 36.7 | 2.417E-89 | 301 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_469373.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14008 | STKc_LKB1_CaMKK | - | 48 | 407 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 42 | 407 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K07359 |
| K21157 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
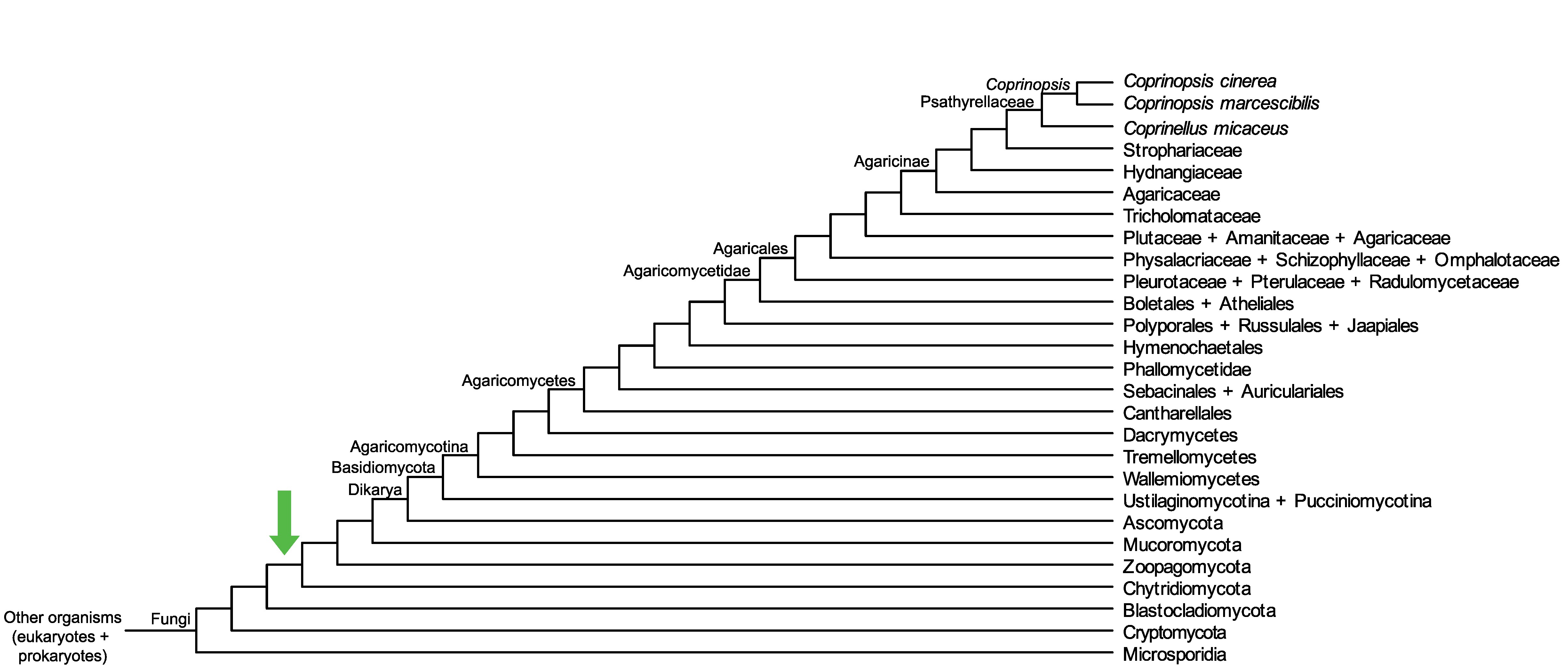
Conservation of CopciAB_469373 across fungi.
Arrow shows the origin of gene family containing CopciAB_469373.
Protein
| Sequence id | CopciAB_469373.T0 |
|---|---|
| Sequence |
>CopciAB_469373.T0 MAQGGAMGYQTPDSLQLETTQPLIFTTRLHTRSSRRKKLNQYVKCERIGKGRHGEVYLCRDGHREVAIKAVKRSN PKDKIKLLRKTYQQQDESGPQKRPKLSSTENSIRKEIAVMKQCRHHPNLAMLYEVIDDPKEEKIYLIMEYLSGGP VEWSTQDDKPLLTLAQTRRVMRDVILGLEFMHARNILHRDIKPSNIMYTEDRRSVKLIDFGVSHIIYPPADKKRK LDADECEAYERSLFTRSDLLKRIGTPSFLAPEVVWFSDTLDLSSSASYDVTLPTSFESPSDTVVGSTFSMPKVRP PITKAVDIWALAVTFYCLLFGHTPFTGSSSSENDNPHHQEFMLYHQICNQDWTIDDVMGAERIPTGGRRPRDATS DGYLVVQLLEGMLQKDPRDRMPLSEVKKNPFILRDVYNAKDWLRLTAQESTESEPCYQWLKVAARKFVNLLSKQA RS |
| Length | 452 |
Coding
| Sequence id | CopciAB_469373.T0 |
|---|---|
| Sequence |
>CopciAB_469373.T0 ATGGCCCAGGGTGGTGCCATGGGATACCAAACCCCTGACTCGTTACAACTCGAAACGACACAACCATTAATCTTC ACTACACGCTTGCACACGCGGTCGAGTCGACGGAAGAAACTCAACCAATACGTCAAATGCGAGAGAATAGGCAAA GGGAGACATGGGGAGGTTTATCTATGTCGGGATGGCCATCGCGAAGTGGCGATTAAAGCTGTCAAACGAAGCAAT CCCAAGGACAAGATTAAGCTACTGAGGAAGACATATCAACAGCAGGATGAATCGGGACCGCAGAAGCGTCCCAAG TTGAGCAGCACGGAGAACAGCATACGGAAAGAGATTGCTGTCATGAAGCAGTGTCGGCACCATCCTAATCTCGCG ATGCTGTACGAAGTCATTGATGATCCAAAGGAGGAAAAGATATACCTTATCATGGAATACCTATCCGGAGGTCCT GTCGAATGGTCGACCCAGGACGACAAGCCATTATTGACCTTGGCGCAGACGCGTCGCGTTATGAGGGACGTCATA CTGGGTCTCGAATTCATGCACGCTAGGAACATCCTGCATCGAGATATCAAGCCCTCCAACATCATGTACACCGAA GACCGCCGGTCTGTCAAGCTGATAGACTTTGGCGTGTCTCATATCATTTACCCACCAGCCGACAAGAAACGAAAG CTGGACGCAGACGAATGTGAAGCCTACGAGAGGTCTTTATTCACCCGGTCTGACTTGTTGAAGCGAATCGGGACC CCTTCATTCCTAGCACCGGAAGTGGTGTGGTTCTCCGACACCCTTGACTTGTCGAGTTCCGCATCCTACGACGTG ACCCTGCCTACGTCTTTCGAGAGCCCGTCCGACACTGTGGTTGGAAGCACCTTCTCAATGCCCAAAGTTCGCCCG CCGATAACCAAAGCTGTGGATATCTGGGCTCTCGCAGTAACTTTCTATTGCCTTCTTTTCGGACACACCCCATTC ACAGGCTCTTCTTCCTCTGAGAACGACAATCCCCATCACCAGGAGTTCATGCTCTACCACCAAATTTGCAACCAA GATTGGACCATTGACGACGTGATGGGGGCCGAGCGGATCCCAACTGGAGGTCGTAGACCAAGGGATGCCACGAGT GATGGATACCTGGTGGTACAGTTGCTCGAAGGGATGCTTCAAAAGGATCCCAGAGATCGCATGCCCCTCTCGGAA GTGAAGAAGAATCCATTCATCCTTCGGGATGTATACAATGCAAAGGACTGGCTTCGCCTAACAGCCCAAGAGTCG ACCGAGTCAGAGCCTTGCTACCAATGGCTCAAGGTTGCCGCCCGAAAATTCGTCAACTTGCTTTCGAAACAAGCT CGCTCATGA |
| Length | 1359 |
Transcript
| Sequence id | CopciAB_469373.T0 |
|---|---|
| Sequence |
>CopciAB_469373.T0 ATGAACTTAAACCCCGACCCCAATGTCCTGACTTGCTTCTTTGCTCCTTTGCAGCCCCTTTACAACTAACGGCCA TGGCCCAGGGTGGTGCCATGGGATACCAAACCCCTGACTCGTTACAACTCGAAACGACACAACCATTAATCTTCA CTACACGCTTGCACACGCGGTCGAGTCGACGGAAGAAACTCAACCAATACGTCAAATGCGAGAGAATAGGCAAAG GGAGACATGGGGAGGTTTATCTATGTCGGGATGGCCATCGCGAAGTGGCGATTAAAGCTGTCAAACGAAGCAATC CCAAGGACAAGATTAAGCTACTGAGGAAGACATATCAACAGCAGGATGAATCGGGACCGCAGAAGCGTCCCAAGT TGAGCAGCACGGAGAACAGCATACGGAAAGAGATTGCTGTCATGAAGCAGTGTCGGCACCATCCTAATCTCGCGA TGCTGTACGAAGTCATTGATGATCCAAAGGAGGAAAAGATATACCTTATCATGGAATACCTATCCGGAGGTCCTG TCGAATGGTCGACCCAGGACGACAAGCCATTATTGACCTTGGCGCAGACGCGTCGCGTTATGAGGGACGTCATAC TGGGTCTCGAATTCATGCACGCTAGGAACATCCTGCATCGAGATATCAAGCCCTCCAACATCATGTACACCGAAG ACCGCCGGTCTGTCAAGCTGATAGACTTTGGCGTGTCTCATATCATTTACCCACCAGCCGACAAGAAACGAAAGC TGGACGCAGACGAATGTGAAGCCTACGAGAGGTCTTTATTCACCCGGTCTGACTTGTTGAAGCGAATCGGGACCC CTTCATTCCTAGCACCGGAAGTGGTGTGGTTCTCCGACACCCTTGACTTGTCGAGTTCCGCATCCTACGACGTGA CCCTGCCTACGTCTTTCGAGAGCCCGTCCGACACTGTGGTTGGAAGCACCTTCTCAATGCCCAAAGTTCGCCCGC CGATAACCAAAGCTGTGGATATCTGGGCTCTCGCAGTAACTTTCTATTGCCTTCTTTTCGGACACACCCCATTCA CAGGCTCTTCTTCCTCTGAGAACGACAATCCCCATCACCAGGAGTTCATGCTCTACCACCAAATTTGCAACCAAG ATTGGACCATTGACGACGTGATGGGGGCCGAGCGGATCCCAACTGGAGGTCGTAGACCAAGGGATGCCACGAGTG ATGGATACCTGGTGGTACAGTTGCTCGAAGGGATGCTTCAAAAGGATCCCAGAGATCGCATGCCCCTCTCGGAAG TGAAGAAGAATCCATTCATCCTTCGGGATGTATACAATGCAAAGGACTGGCTTCGCCTAACAGCCCAAGAGTCGA CCGAGTCAGAGCCTTGCTACCAATGGCTCAAGGTTGCCGCCCGAAAATTCGTCAACTTGCTTTCGAAACAAGCTC GCTCATGACACCTCGTTTTTCATCTGCCGGTGTATTGTATCTATTATCTGTTGCTTATCTTCGAACATTTTCTGC TGGGCCTCTCATGTATTAGTCGTACTCTATCTCTACGTCTACGACTCGCCTTTC |
| Length | 1554 |
Gene
| Sequence id | CopciAB_469373.T0 |
|---|---|
| Sequence |
>CopciAB_469373.T0 ATGAACTTAAACCCCGACCCCAATGTCCTGACTTGCTTCTTTGCTCCTTTGCAGCCCCTTTACAACTAACGGCCA TGGCCCAGGGTGGTGCCATGGGATACCAAACCCCTGACTCGTTACAACTCGAAACGACACAACCATTAATCTTCA CTACACGCTTGCACACGCGGTCGAGTCGACGGAAGAAACTCAACCAATACGTCAAATGCGAGAGAATAGGCAAAG GGAGACATGGGGAGGTTTATCTATGTCGGGATGGCCATCGCGAAGTGGTGCGTTGTGAAGCGCCCAGGATTCCTG TGGTCTTTTGCGCTCATTCTAGTCTCGGTGTTTAGGCGATTAAAGCTGTCAAACGAAGCAATCCCAAGGACAAGA TTAAGCTACTGAGGAAGACATATCAACAGCAGGATGAATCGGGACCGCAGAAGCGTCCCAAGTTGAGCAGCACGG AGAACAGCATACGGAAAGAGATTGCTGTCATGAAGCAGTGTCGGCACCATCCTAATCTCGCGATGCTGTACGAAG TCATTGATGATCCAAAGGAGGAAAAGATATACCTTAGTGGGTGGCAGATGAACTGTAGGGCGTTGGTCGTATTCT GAATCACTTGCAGTCATGGAATACCTATCCGGAGGTCCTGTCGAATGGTCGACCCAGGACGACAAGCCATTATTG ACCTTGGCGCAGACGCGTCGCGTTATGAGGGACGTCATACTGGGTCTCGAATTCAGTTCGTGGTCTCCCTGTTGG TATCGTGGTTGTGTTCTAACTAGTTTTCAGTGCACGCTAGGAACATCCTGCATCGAGATATCAAGCCCTCCAACA TCATGTACACCGAAGACCGCCGGTCTGTCAAGCTGATAGACTTTGGCGTGTCTCATATCATTTACCCACCAGCCG ACAAGAAACGAAAGCTGGACGCAGACGAATGTGAAGCCTACGAGAGGTCTTTATTCACCCGGTCTGACTTGTTGA AGCGAATCGGGACCCCTTCATTCCTAGCACCGGAAGTGGTGTGGTTCTCCGACACCCTTGACTTGTCGAGTTCCG CATCCTACGACGTGACCCTGCCTACGTCTTTCGAGAGCCCGTCCGACACTGTGGTTGGAAGCACCTTCTCAATGC CCAAAGTTCGCCCGCCGATAACCAAAGCTGTGGATATCTGGGCTCTCGCAGTAACTTTCTATTGCCTTCTTTTCG GACACACCCCATTCACAGGCTCTTCTTCCTCTGAGAACGACAATCCCCATCACCAGGAGTTCATGCTCTACCACC AAATTTGCAACCAAGATTGGACCATTGACGACGTGATGGGGGCCGAGCGGATCCCAACTGGAGGTCGTAGACCAA GGGATGCCACGAGTGATGGATACCTGGTGGTACAGTTGCTCGAAGGGATGCTTCAAAAGGATCCCAGAGATCGCA TGCCCCTCTCGGAAGTGAAGGTACGAAAGTCGATACTGATATAAACTTGTTCATTGCCCCTAACCGCCTCTACAG AAGAATCCATTCATCCTTCGGGATGTATACAATGCAAAGGACTGGCTTCGCCTAACAGCCCAAGAGTCGACCGAG TCAGAGCCTTGCTACCAATGGCTCAAGGTTGCCGCCCGAAAATTCGTCAACTTGCTTTCGAAACAAGCTCGCTCA TGACACCTCGTTTTTCATCTGCCGGTGTATTGTATCTATTATCTGTTGCTTATCTTCGAACATTTTCTGCTGGGC CTCTCATGTATTAGTCGTACTCTATCTCTACGTCTACGACTCGCCTTTC |
| Length | 1774 |