CopciAB_469725
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_469725 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | BUB1 | Synonyms | 469725 |
| Uniprot id | Functional description | Mad3/BUB1 hoMad3/BUB1 homology region 1 | |
| Location | scaffold_8:642875..644527 | Strand | + |
| Gene length (nt) | 1653 | Transcript length (nt) | 1532 |
| CDS length (nt) | 1407 | Protein length (aa) | 468 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | mad3 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7267225 | 62.6 | 9.095E-182 | 570 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29894 | 59.6 | 8.145E-166 | 524 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_152609 | 59.1 | 6.062E-164 | 518 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_122130 | 58.7 | 3.372E-162 | 513 |
| Lentinula edodes NBRC 111202 | Lenedo1_1090107 | 57.6 | 3.539E-155 | 493 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_9089 | 57.5 | 3.511E-155 | 493 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1440200 | 54.2 | 1.099E-150 | 480 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1074434 | 54 | 2.068E-150 | 479 |
| Pleurotus ostreatus PC9 | PleosPC9_1_100738 | 54 | 2.024E-150 | 479 |
| Flammulina velutipes | Flave_chr10AA01105 | 54.9 | 1.111E-139 | 448 |
| Schizophyllum commune H4-8 | Schco3_2636410 | 51.2 | 1.077E-132 | 428 |
| Lentinula edodes B17 | Lened_B_1_1_9515 | 50.7 | 1.296E-125 | 407 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_75678 | 44 | 2.757E-117 | 383 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | BUB1 |
|---|---|
| Protein id | CopciAB_469725.T0 |
| Description | Mad3/BUB1 hoMad3/BUB1 homology region 1 |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF08311 | Mad3/BUB1 homology region 1 | IPR013212 | 63 | 185 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR015661 | Mitotic spindle checkpoint protein Bub1/Mad3 |
| IPR013212 | Mad3/Bub1 homology region 1 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0007094 | mitotic spindle assembly checkpoint | BP |
KEGG
| KEGG Orthology |
|---|
| K02178 |
| K06680 |
EggNOG
| COG category | Description |
|---|---|
| D | Mad3/BUB1 hoMad3/BUB1 homology region 1 |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
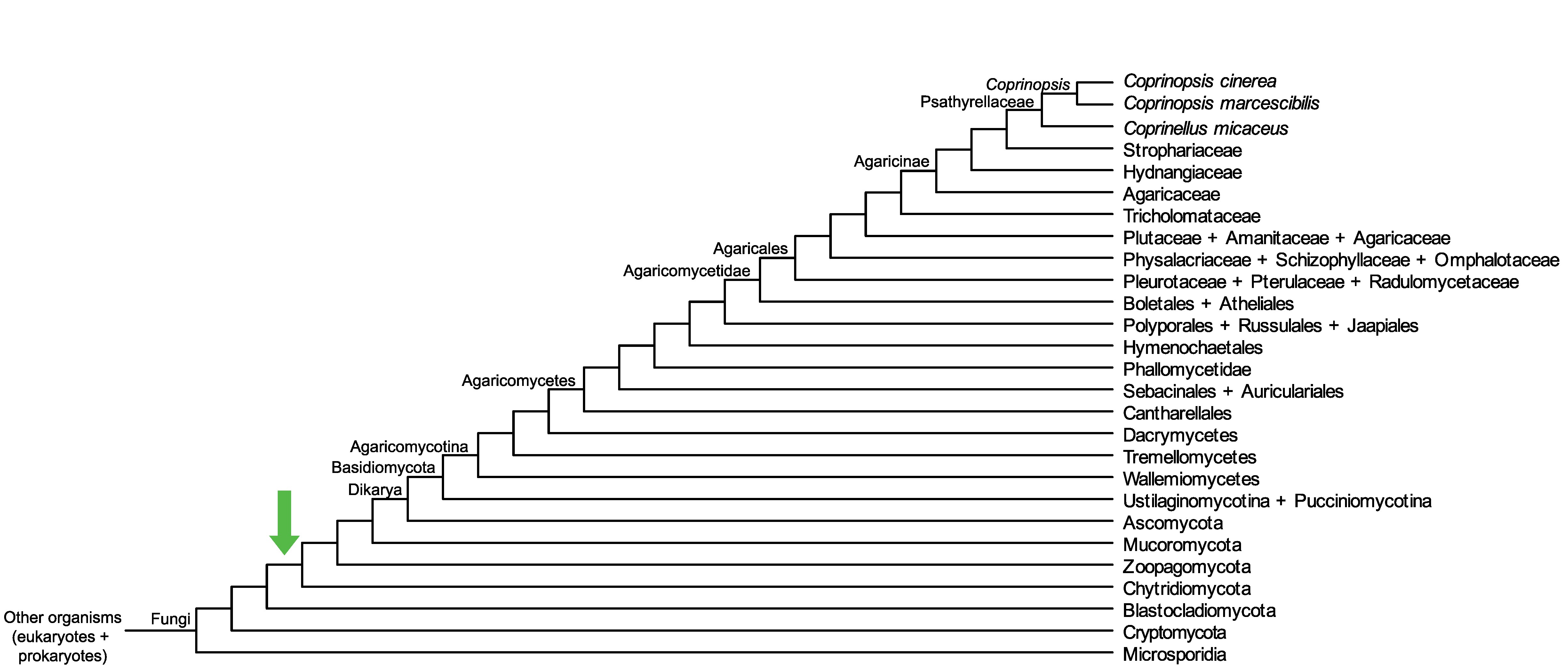
Conservation of CopciAB_469725 across fungi.
Arrow shows the origin of gene family containing CopciAB_469725.
Protein
| Sequence id | CopciAB_469725.T0 |
|---|---|
| Sequence |
>CopciAB_469725.T0 MISADDVFQESEPVVVDCDLLEAAKENVQPLATGRRVTALSTVLATPAAQRDAKLSATRNRLRINVELALEDEDG DPLDAYCRLVYWTMENYPQGHSAESGLLELLEEATRVLKDHNGGVYRGDMKYLKLWLIYASYVERPTIIYRFLLA NDIGIGFALLYEEHAAVLERDGRKQEADEIYALGIARKAEPLDHLESRYQDFQKRMISTFTAPAPEPARATQAAQ PKRQALATTSKIPTSTSRTGGTRIASSSRNTTVAAPTNMPPPSNRGGFQIFVDPSGQNGEAGEGGENGWHDLGTR KTRVKENVPETKKLVGTTIKQAGKTRRVVSASAASGSGSGSSGSSGSKFAIFRDPVDAPTPTPPAPAAASSSRKP SSSGKATKAFVPFVDEADVKKPSPTPPETPRAATPKFTPFRDEGDAESSRATAVPVGDAVIKVKKTGPKAPVPTS EAEALRKDPLKNYSTNDT |
| Length | 468 |
Coding
| Sequence id | CopciAB_469725.T0 |
|---|---|
| Sequence |
>CopciAB_469725.T0 ATGATATCCGCAGACGACGTTTTCCAAGAATCTGAGCCCGTCGTCGTCGACTGCGACCTTCTCGAGGCTGCCAAA GAGAATGTTCAGCCCCTCGCCACCGGTCGACGTGTGACGGCTCTTTCCACTGTTCTGGCGACCCCCGCCGCACAA CGCGATGCTAAATTGTCGGCGACACGAAATAGACTTCGAATCAACGTCGAGCTAGCACTGGAAGATGAAGACGGT GATCCATTGGACGCTTACTGTCGTCTCGTGTACTGGACCATGGAGAACTACCCTCAAGGTCACAGCGCAGAGTCG GGGCTGTTGGAACTGTTGGAAGAAGCCACTCGCGTTCTCAAAGACCACAACGGCGGGGTGTACCGTGGAGACATG AAGTACCTCAAGTTATGGCTGATATACGCCAGCTACGTGGAACGGCCCACCATCATTTATCGGTTCCTGCTCGCC AATGATATCGGAATCGGGTTCGCGCTACTGTACGAAGAGCACGCAGCTGTTCTAGAAAGGGATGGCCGGAAACAA GAGGCAGATGAGATTTATGCTTTGGGTATCGCTCGTAAAGCAGAACCCCTCGATCACCTAGAAAGCCGGTATCAA GATTTCCAGAAGAGGATGATCTCGACCTTTACGGCGCCTGCCCCTGAACCCGCGAGGGCGACCCAAGCTGCTCAG CCTAAACGTCAGGCGCTCGCTACGACGTCGAAGATCCCCACCTCGACATCGAGAACCGGCGGCACGCGAATCGCG TCATCGAGCCGAAATACGACGGTAGCTGCTCCCACCAATATGCCACCACCCTCGAATCGTGGGGGATTCCAAATC TTTGTGGATCCTTCTGGTCAGAATGGAGAAGCGGGTGAAGGAGGGGAGAATGGGTGGCACGATCTCGGAACACGT AAGACGAGGGTGAAGGAGAATGTTCCGGAGACGAAGAAACTCGTTGGGACGACCATCAAGCAGGCTGGGAAGACG AGGAGGGTGGTATCGGCTTCGGCTGCTTCTGGTTCTGGGTCCGGCTCCTCGGGAAGCTCAGGCTCCAAGTTTGCG ATATTCAGGGACCCAGTGGATGCCCCAACCCCAACCCCACCAGCGCCTGCTGCGGCTTCATCCTCGAGAAAGCCG TCGTCTAGCGGGAAGGCTACCAAGGCATTTGTGCCTTTTGTTGATGAGGCTGATGTGAAAAAGCCATCTCCTACA CCACCGGAGACTCCTCGTGCTGCGACGCCTAAGTTCACTCCCTTCCGTGACGAGGGTGATGCTGAGTCATCGCGG GCGACTGCAGTCCCAGTGGGCGATGCAGTGATCAAGGTCAAGAAGACAGGGCCCAAGGCTCCGGTGCCAACTTCT GAAGCAGAGGCGCTCAGAAAAGATCCTCTGAAAAACTATTCGACGAATGATACGTGA |
| Length | 1407 |
Transcript
| Sequence id | CopciAB_469725.T0 |
|---|---|
| Sequence |
>CopciAB_469725.T0 GAACAACTTTAGTCTTTGACCTCGACGACCATGATATCCGCAGACGACGTTTTCCAAGAATCTGAGCCCGTCGTC GTCGACTGCGACCTTCTCGAGGCTGCCAAAGAGAATGTTCAGCCCCTCGCCACCGGTCGACGTGTGACGGCTCTT TCCACTGTTCTGGCGACCCCCGCCGCACAACGCGATGCTAAATTGTCGGCGACACGAAATAGACTTCGAATCAAC GTCGAGCTAGCACTGGAAGATGAAGACGGTGATCCATTGGACGCTTACTGTCGTCTCGTGTACTGGACCATGGAG AACTACCCTCAAGGTCACAGCGCAGAGTCGGGGCTGTTGGAACTGTTGGAAGAAGCCACTCGCGTTCTCAAAGAC CACAACGGCGGGGTGTACCGTGGAGACATGAAGTACCTCAAGTTATGGCTGATATACGCCAGCTACGTGGAACGG CCCACCATCATTTATCGGTTCCTGCTCGCCAATGATATCGGAATCGGGTTCGCGCTACTGTACGAAGAGCACGCA GCTGTTCTAGAAAGGGATGGCCGGAAACAAGAGGCAGATGAGATTTATGCTTTGGGTATCGCTCGTAAAGCAGAA CCCCTCGATCACCTAGAAAGCCGGTATCAAGATTTCCAGAAGAGGATGATCTCGACCTTTACGGCGCCTGCCCCT GAACCCGCGAGGGCGACCCAAGCTGCTCAGCCTAAACGTCAGGCGCTCGCTACGACGTCGAAGATCCCCACCTCG ACATCGAGAACCGGCGGCACGCGAATCGCGTCATCGAGCCGAAATACGACGGTAGCTGCTCCCACCAATATGCCA CCACCCTCGAATCGTGGGGGATTCCAAATCTTTGTGGATCCTTCTGGTCAGAATGGAGAAGCGGGTGAAGGAGGG GAGAATGGGTGGCACGATCTCGGAACACGTAAGACGAGGGTGAAGGAGAATGTTCCGGAGACGAAGAAACTCGTT GGGACGACCATCAAGCAGGCTGGGAAGACGAGGAGGGTGGTATCGGCTTCGGCTGCTTCTGGTTCTGGGTCCGGC TCCTCGGGAAGCTCAGGCTCCAAGTTTGCGATATTCAGGGACCCAGTGGATGCCCCAACCCCAACCCCACCAGCG CCTGCTGCGGCTTCATCCTCGAGAAAGCCGTCGTCTAGCGGGAAGGCTACCAAGGCATTTGTGCCTTTTGTTGAT GAGGCTGATGTGAAAAAGCCATCTCCTACACCACCGGAGACTCCTCGTGCTGCGACGCCTAAGTTCACTCCCTTC CGTGACGAGGGTGATGCTGAGTCATCGCGGGCGACTGCAGTCCCAGTGGGCGATGCAGTGATCAAGGTCAAGAAG ACAGGGCCCAAGGCTCCGGTGCCAACTTCTGAAGCAGAGGCGCTCAGAAAAGATCCTCTGAAAAACTATTCGACG AATGATACGTGAACTCGCCTATACCTATTCTTGCACATATGTCCTTTCCTTTTTTGCACGGTGTATTGTCATTTA CTCATTGCTTGAATTCCATTGTATTTGCTGCA |
| Length | 1532 |
Gene
| Sequence id | CopciAB_469725.T0 |
|---|---|
| Sequence |
>CopciAB_469725.T0 GAACAACTTTAGTCTTTGACCTCGACGACCATGATATCCGCAGACGACGTTTTCCAAGAATCTGAGCCCGTCGTC GTCGACTGCGACCTTCTCGAGGCTGCCAAAGAGAATGTTCAGCCCCTCGCCACCGGTCGACGTGTGACGGCTCTT TCCACTGTTCTGGCGACCCCCGCCGCACAACGCGATGCTAAATTGTCGGCGACACGAAATAGACTTCGAATCAAC GTCGAGCTAGCACTGGAAGATGAAGACGGTGATCCATTGGACGCTTACTGTCGTCTCGTGTACTGGACCATGGAG AACTACCCTCAAGGTCACAGCGCAGAGTCGGGGCTGTTGGAACTGTTGGAAGAAGCCACTCGCGTTCTCAAAGAC CACAACGGCGGGGTGTACCGTGGAGACATGAAGTACCTCAAGTTATGGCTGATATACGCCAGCTACGTGGAACGG CCCACCATCATTTATCGGTTCCTGCTCGCCAATGATATCGGAATCGGGTTCGCGCTACTGTACGAAGAGCACGCA GCTGTTCTAGAAAGGGATGGCCGGTGAGTACGAATGTATACCTCTATCCCATGGTTACGCTTTGAGCTCAATCAT TGCGAGAACTCAGGAAACAAGAGGCAGATGAGATTTATGCTTTGGGTATCGCTCGTAAAGCAGAACCCCTCGATC ACCTAGAAAGCCGGTATCAAGATTTCCAGAAGAGGATGATCTCGACCTTTACGGCGCCTGCCCCTGAACCCGCGA GGGCGACCCAAGCTGCTCAGCCTAAACGTCAGGCGCTCGCTACGACGTCGAAGATCCCCACCTCGACATCGAGAA CCGGCGGCACGCGAATCGCGTCATCGAGCCGAAATACGACGGTAGCTGCTCCCACCAATATGCCACCACCCTCGA ATCGTGGGGGATTCCAAATCTTTGTGGATCCTTCTGGTCAGAATGGAGAAGCGGGTGAAGGAGGGGAGAATGGGT GGCACGATCTCGGAACACGTAAGACGAGGGTGAAGGAGAATGTTCCGGAGACGAAGAAACTCGTTGGGACGACCA TCAAGCAGGCTGGGAAGACGAGGAGGGTGGTATCGGCTTCGGCTGCTTCTGGTTCTGGGTCCGGCTCCTCGGGAA GCTCAGGCTCCAAGTTTGCGATATTCAGGGACCCAGTGGATGCCCCAACCCCAACCCCACCAGCGCCTGCTGCGG CTTCATCCTCGAGAAAGCCGTCGTCTAGCGGGAAGGCTACCAAGGCATTTGTGCCTTTTGTTGATGAGGCTGATG TGAAAAAGCCATCTCCTACACCACCGGAGACTCCTCGTGCTGCGACGCCTAAGTTCACTCCCTTCCGTGACGAGG TAAGCTCTCCATAACTCTCCTTCATCGATAACCTGACTGATCGCCTTATCCTTAGGGTGATGCTGAGTCATCGCG GGCGACTGCAGTCCCAGTGGGCGATGCAGTGATCAAGGTCAAGAAGACAGGGCCCAAGGCTCCGGTGCCAACTTC TGAAGCAGAGGCGCTCAGAAAAGATCCTCTGAAAAACTATTCGACGAATGATACGTGAACTCGCCTATACCTATT CTTGCACATATGTCCTTTCCTTTTTTGCACGGTGTATTGTCATTTACTCATTGCTTGAATTCCATTGTATTTGCT GCA |
| Length | 1653 |