CopciAB_470611
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_470611 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | cmk1 | Synonyms | 470611 |
| Uniprot id | Functional description | Belongs to the protein kinase superfamily | |
| Location | scaffold_9:1539249..1541744 | Strand | - |
| Gene length (nt) | 2496 | Transcript length (nt) | 1707 |
| CDS length (nt) | 1467 | Protein length (aa) | 488 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | cmk1 |
| Aspergillus nidulans | AN3065_cmkB |
| Neurospora crassa | camk-2 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7261890 | 62.7 | 1.307E-196 | 614 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28099 | 72 | 7.234E-196 | 612 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1060462 | 80.1 | 5.268E-196 | 612 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1420414 | 65.2 | 1.498E-194 | 608 |
| Pleurotus ostreatus PC9 | PleosPC9_1_114537 | 79.6 | 3.012E-193 | 604 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_194968 | 69.4 | 6.997E-191 | 597 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114778 | 69.2 | 7.292E-191 | 597 |
| Schizophyllum commune H4-8 | Schco3_2668685 | 63.6 | 1.073E-190 | 597 |
| Lentinula edodes B17 | Lened_B_1_1_14952 | 72.3 | 5.18E-186 | 583 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_22270 | 72.3 | 6.496E-186 | 583 |
| Lentinula edodes NBRC 111202 | Lenedo1_1139054 | 75.8 | 9.204E-182 | 571 |
| Auricularia subglabra | Aurde3_1_1272285 | 64.1 | 3.596E-180 | 567 |
| Flammulina velutipes | Flave_chr11AA01396 | 73.3 | 1.193E-177 | 559 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_130823 | 74.6 | 1.29E-176 | 556 |
| Grifola frondosa | Grifr_OBZ76298 | 67.4 | 1.289E-176 | 556 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | cmk1 |
|---|---|
| Protein id | CopciAB_470611.T0 |
| Description | Belongs to the protein kinase superfamily |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd05117 | STKc_CAMK | - | 106 | 367 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 107 | 368 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR017441 | Protein kinase, ATP binding site |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08794 |
EggNOG
| COG category | Description |
|---|---|
| T | Belongs to the protein kinase superfamily |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
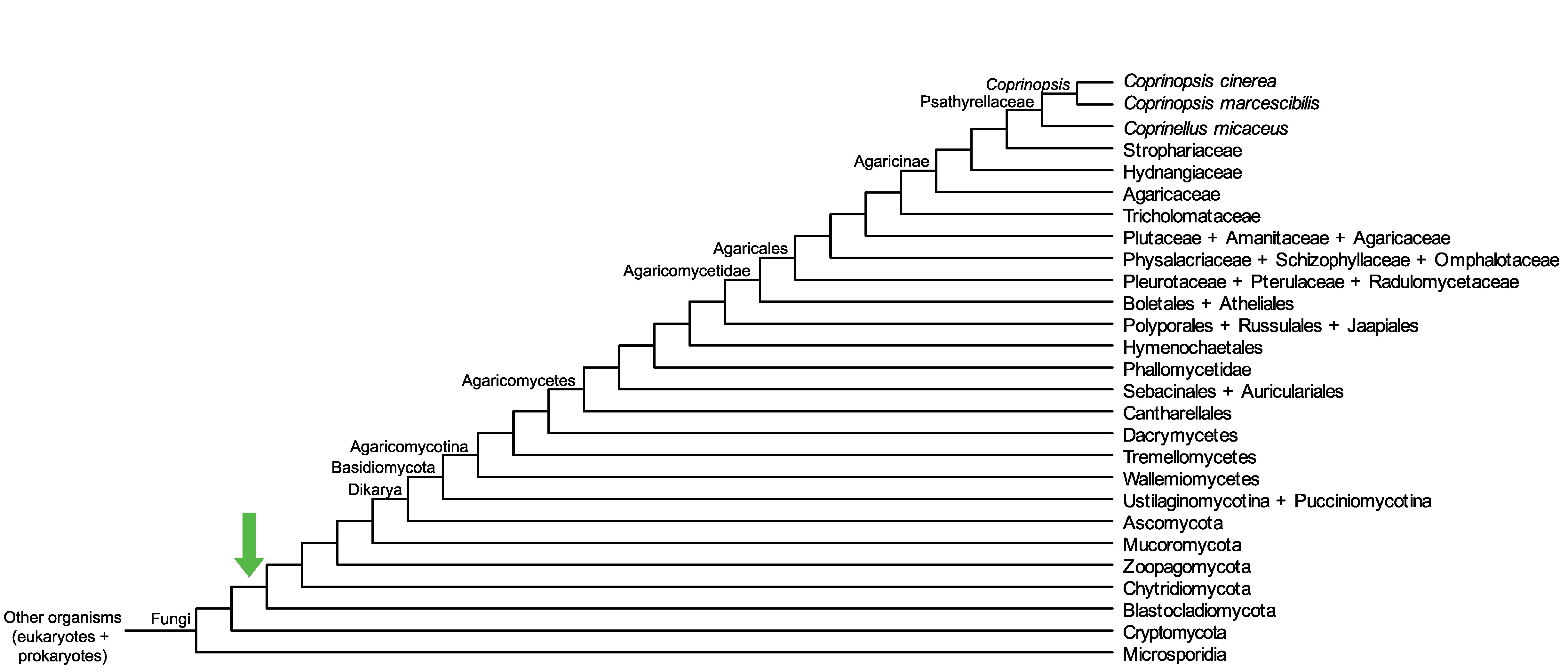
Conservation of CopciAB_470611 across fungi.
Arrow shows the origin of gene family containing CopciAB_470611.
Protein
| Sequence id | CopciAB_470611.T0 |
|---|---|
| Sequence |
>CopciAB_470611.T0 MGSAQSKQDGNPDRLKRISAAFKKPRSSEESTRPQAPPPPPVAAPAAQVSDAGSANESHLKKGGDNDERAVMHDS VQRNAPHPALKQPQQPQNQQGQPMPATVPCQYRTGKTLGSGTYAIVKEAIHITTGKYYACKVINKKLMEGREHMV RNEIAVLKKISSGHPNIVTLHDYFETAHNLYLVFDLCTGGELFDRICAKGNYYEADAADLVRTIFKAVEYIHNSG VVHRDLKPENLLFKTQAEEAAIMIADFGLSRVMEEEKFSLLTEICGTPGYMAPEIFKRTGHGKPVDVWAMGVITY FLLAGYTPFDRESQQQEMEAIIAGDYKFEPAEYWENVSETAKDFVRTCLTIDPNKRPTATEALAHKWLADKEPHF VANPADGTPADLLPNVKRAFNAKRAWKKAAYGIRAMNRMASLASGGLTNPEAALMRENLDKYKAESEKEDIQTAS ITYMPQHTPVTAEFPAGSQKSATDDELQKKLANATIQG |
| Length | 488 |
Coding
| Sequence id | CopciAB_470611.T0 |
|---|---|
| Sequence |
>CopciAB_470611.T0 ATGGGCTCTGCTCAAAGTAAACAAGACGGAAACCCTGACCGCCTCAAACGCATCTCTGCCGCGTTCAAGAAACCT CGCAGCTCTGAAGAGTCTACCAGACCACAAGCACCACCACCACCACCAGTGGCAGCACCAGCAGCCCAAGTCTCA GACGCAGGCAGCGCCAACGAGTCGCATTTGAAAAAGGGCGGCGACAACGATGAGCGGGCAGTGATGCACGATTCA GTGCAACGCAACGCGCCTCATCCCGCTCTCAAACAACCACAACAACCACAGAACCAACAAGGCCAACCAATGCCA GCTACAGTCCCCTGCCAGTATCGCACCGGCAAGACATTGGGCAGCGGAACATACGCTATCGTCAAGGAGGCCATC CACATTACAACCGGAAAGTACTATGCCTGCAAAGTTATCAACAAGAAACTTATGGAGGGCCGTGAACACATGGTC CGTAACGAGATCGCCGTCCTAAAGAAGATATCCAGTGGTCACCCCAACATTGTAACCCTCCACGATTACTTTGAG ACCGCCCACAATCTCTACCTCGTCTTCGATCTCTGCACCGGTGGTGAACTCTTCGACCGGATATGCGCAAAGGGC AACTACTACGAGGCCGATGCAGCCGACCTGGTCCGCACCATCTTCAAAGCCGTTGAATACATCCACAACTCTGGC GTCGTTCATCGAGACCTGAAACCCGAGAACCTCCTCTTCAAGACCCAAGCCGAAGAAGCAGCCATCATGATCGCA GACTTTGGATTGAGTAGGGTCATGGAAGAAGAAAAGTTTAGCCTCTTGACCGAGATCTGCGGTACACCCGGTTAC ATGGCACCGGAAATCTTCAAACGAACCGGCCATGGCAAACCGGTTGACGTCTGGGCAATGGGCGTCATCACGTAC TTCCTCCTCGCGGGCTACACGCCCTTTGATCGTGAATCCCAGCAACAAGAAATGGAAGCCATCATCGCCGGAGAC TACAAATTCGAACCAGCAGAATATTGGGAGAACGTATCCGAAACCGCCAAGGACTTTGTGAGGACGTGTTTGACC ATCGACCCCAACAAGAGGCCGACAGCGACGGAGGCGCTGGCTCACAAGTGGCTGGCGGACAAAGAACCTCACTTT GTTGCTAACCCTGCTGACGGTACTCCAGCTGACCTCTTGCCAAACGTTAAACGAGCATTCAACGCCAAGCGAGCC TGGAAAAAGGCGGCGTACGGTATCCGCGCGATGAACAGGATGGCGTCGCTCGCCAGCGGAGGATTGACGAACCCA GAGGCAGCACTCATGCGCGAGAACCTGGACAAGTACAAGGCAGAGTCAGAGAAGGAGGATATTCAGACCGCATCG ATCACATACATGCCACAACACACGCCTGTCACAGCCGAGTTCCCAGCAGGTTCTCAAAAGAGCGCAACCGACGAC GAATTGCAGAAGAAACTCGCCAACGCCACCATCCAGGGTTAA |
| Length | 1467 |
Transcript
| Sequence id | CopciAB_470611.T0 |
|---|---|
| Sequence |
>CopciAB_470611.T0 AGGGTCCTTTACTCGCTTCCTCACTTTCCCCCAACTCTTTCTCAAATGGGCTCTGCTCAAAGTAAACAAGACGGA AACCCTGACCGCCTCAAACGCATCTCTGCCGCGTTCAAGAAACCTCGCAGCTCTGAAGAGTCTACCAGACCACAA GCACCACCACCACCACCAGTGGCAGCACCAGCAGCCCAAGTCTCAGACGCAGGCAGCGCCAACGAGTCGCATTTG AAAAAGGGCGGCGACAACGATGAGCGGGCAGTGATGCACGATTCAGTGCAACGCAACGCGCCTCATCCCGCTCTC AAACAACCACAACAACCACAGAACCAACAAGGCCAACCAATGCCAGCTACAGTCCCCTGCCAGTATCGCACCGGC AAGACATTGGGCAGCGGAACATACGCTATCGTCAAGGAGGCCATCCACATTACAACCGGAAAGTACTATGCCTGC AAAGTTATCAACAAGAAACTTATGGAGGGCCGTGAACACATGGTCCGTAACGAGATCGCCGTCCTAAAGAAGATA TCCAGTGGTCACCCCAACATTGTAACCCTCCACGATTACTTTGAGACCGCCCACAATCTCTACCTCGTCTTCGAT CTCTGCACCGGTGGTGAACTCTTCGACCGGATATGCGCAAAGGGCAACTACTACGAGGCCGATGCAGCCGACCTG GTCCGCACCATCTTCAAAGCCGTTGAATACATCCACAACTCTGGCGTCGTTCATCGAGACCTGAAACCCGAGAAC CTCCTCTTCAAGACCCAAGCCGAAGAAGCAGCCATCATGATCGCAGACTTTGGATTGAGTAGGGTCATGGAAGAA GAAAAGTTTAGCCTCTTGACCGAGATCTGCGGTACACCCGGTTACATGGCACCGGAAATCTTCAAACGAACCGGC CATGGCAAACCGGTTGACGTCTGGGCAATGGGCGTCATCACGTACTTCCTCCTCGCGGGCTACACGCCCTTTGAT CGTGAATCCCAGCAACAAGAAATGGAAGCCATCATCGCCGGAGACTACAAATTCGAACCAGCAGAATATTGGGAG AACGTATCCGAAACCGCCAAGGACTTTGTGAGGACGTGTTTGACCATCGACCCCAACAAGAGGCCGACAGCGACG GAGGCGCTGGCTCACAAGTGGCTGGCGGACAAAGAACCTCACTTTGTTGCTAACCCTGCTGACGGTACTCCAGCT GACCTCTTGCCAAACGTTAAACGAGCATTCAACGCCAAGCGAGCCTGGAAAAAGGCGGCGTACGGTATCCGCGCG ATGAACAGGATGGCGTCGCTCGCCAGCGGAGGATTGACGAACCCAGAGGCAGCACTCATGCGCGAGAACCTGGAC AAGTACAAGGCAGAGTCAGAGAAGGAGGATATTCAGACCGCATCGATCACATACATGCCACAACACACGCCTGTC ACAGCCGAGTTCCCAGCAGGTTCTCAAAAGAGCGCAACCGACGACGAATTGCAGAAGAAACTCGCCAACGCCACC ATCCAGGGTTAAGCCACGCCCTCTCGTCTACATCCTCATGCCTATGCCTTTCGTCAGTTTGCTTATCCTCGGGCT GTCGTCAATCGGAGTTTACGATCTACCTCTGGCCATCTACCTATCTAATGCTTCGTGATTCATTCCCTTGCTACC ATTGTAGAAATGTATAATGTTGAATATATCGTATTGCCGGGACTAGGACCATTTTCG |
| Length | 1707 |
Gene
| Sequence id | CopciAB_470611.T0 |
|---|---|
| Sequence |
>CopciAB_470611.T0 AGGGTCCTTTACTCGCTTCCTCACTTTCCCCCAACTCTTTCTCAAATGGGCTCTGCTCAAAGTAAACAAGACGGA AACCCTGACCGCCTCAAACGCATCTCTGCCGCGTTCAAGAAACCTCGCAGCTCTGAAGAGTCTACCAGACCACAA GCACCACCACCACCACCAGTGGCAGCACCAGCAGCCCAAGTCTCAGACGCAGGCAGCGCCAACGAGTCGCATTTG AAAAAGGGCGGCGACAACGATGAGCGGGCAGTGATGCACGATTCAGTGCAACGCAACGCGGTGAGGACTGTGTGG CTGTGTCGTTATCACCCGCAAACCATCTGATTACTGGGGGACTCAGCCTCATCCCGCTCTCAAACAACCACAACA ACCACAGAACCAACAAGGCCAACCAATGCCAGCTACAGTCCCCTGCCAGGTCTGTGGTAAACAGACAACATACTC TCTCCTTCCCAATGTTCTAAACACACACGCAGTATCGCACCGGCAAGACATTGGGCAGCGGAACATACGCTATCG TCAAGGAGGCCATCCACATTACAACCGGAAAGTACTATGCCTGCAAAGTTATCAACAAGAAACTTATGGAGGGCC GTGAACACATGGTACCTCTATATCCACTTCGTCTCCAAAAAACGCGTCTCATCCCACTCGCCCTCCAGGTCCGTA ACGAGATCGCCGTCCTAAAGAAGATATCCAGTGGTCACCCCAACATTGTAACCCTCCACGATTACTTTGAGGTTA GCCCTTCACTTTCTCCTTATTCGCCTCTCAACTCTTCTCTTCAGACCGCCCACAATCTCTACCTCGTCTTCGATC TCTGCACCGGTGGTGAACTCTTCGACCGGATATGCGCAAAGGGCAACTACTACGAGGCGTATGTACTCTCATTAA CGTCCATCACACCAATCCTTATCCCATCTACAGCGATGCAGCCGACCTGGTCCGCACCATCTTCAAAGCCGTTGA ATACATCCACAACTCTGGCGTCGTTCATCGAGGTATGCTCTCCCATGTCTTCCACATGCTGCGTGACTCACTCGC TGTCATCTATAATCATTCTTATCATCGTTACTACCTCCAAGACCTGAAACCCGAGAACCTCCTCTTCAAGACCCA AGCCGAAGAAGCAGCCATCATGATCGCAGACTTTGGATTGAGTAGGGTCATGGAAGAAGAAAAGTTTAGCCTCTT GACCGAGATCTGCGGTACACCCGGTGTGAGTTTAATCATCCTCCCTCTCCTATGCTTAAGCTAATCTTCATTGGA ACAGTACATGGCACCGGAAATCTTCAAACGAAGTACGCCCTCTCCCAACTCACATCTCCGATATTATTAACATTA CATTCGCCCAGCCGGCCATGGCAAACCGGTTGACGTCTGGGCAATGGGCGTCATCACGTACTTCCTCCTCGCGGG CTACACGCCCTTTGATCGTGAATCCCAGCAACAAGAAATGGAAGCCATCATCGCCGGAGACTACAAATTCGAACC AGCAGAATATTGGGAGAACGTATCCGAAACCGCCAAGGACTTTGTGAGGACGTGTTTGACCATCGACCCCAACAA GAGGCCGACAGCGACGGAGGCGCTGGCTCACAAGGTGCGCCTATTCGAGTCGTAATGACATTCCCATCCATTCGT GCCGTTGTAACTAATCCATGGATCGCGGTAGTGGCTGGCGGACAAAGAACCTCACTTTGTTGCTAACCCTGCTGA CGGTACTCCAGCTGACCTCTTGCCAAACGTTAAACGAGCATTCAACGCCAAGCGAGCCTGTCCGTATCACCTCCC CATCTTCTATCTCCCATCTTCCTTCTTTAATCTTCATCTTCCCTCCCATCTGCCTCCCATCTATTTATAACGCGT CTATCTTAACCCCGGATCTACGTTTAATTTTCTCAACACCACCCCACTTCGCTCTCTTTTACGACTAACATACTC AAAAACACCCTACTTACCCCTCGCGTACAGGGAAAAAGGCGGCGTACGGTATCCGCGCGATGAACAGGATGGCGT CGCTCGCCAGCGGAGGATTGACGAACCCAGAGGCAGCACTCATGCGCGAGAACCTGGACAAGTACAAGGCAGAGT CAGAGAAGGAGGATATTCAGGTGAGCTGCCTATAAAGATATGTCGCCTGCTTGGTATTTACCATCTTCCCACCAG ACCGCATCGATCACATACATGCCACAACACACGCCTGTCACAGCCGAGTTCCCAGCAGGTTCTCAAAAGAGCGCA ACCGACGACGAATTGCAGAAGAAACTCGCCAACGCCACCATCCAGGGTTAAGCCACGCCCTCTCGTCTACATCCT CATGCCTATGCCTTTCGTCAGTTTGCTTATCCTCGGGCTGTCGTCAATCGGAGTTTACGATCTACCTCTGGCCAT CTACCTATCTAATGCTTCGTGATTCATTCCCTTGCTACCATTGTAGAAATGTATAATGTTGAATATATCGTATTG CCGGGACTAGGACCATTTTCG |
| Length | 2496 |