CopciAB_470894
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_470894 | Strain | |
|---|---|---|---|
| Standard name | - | Synonyms | 470894 |
| Uniprot id | Functional description | Gmc oxidoreductase | |
| Location | scaffold_6:2627001..2629699 | Strand | - |
| Gene length (nt) | 2699 | Transcript length (nt) | 2486 |
| CDS length (nt) | 2199 | Protein length (aa) | 732 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_470894.T0 |
| Description | Gmc oxidoreductase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 62 | 405 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 536 | 673 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR027424 | Glucose Oxidase, domain 2 |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Gmc oxidoreductase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
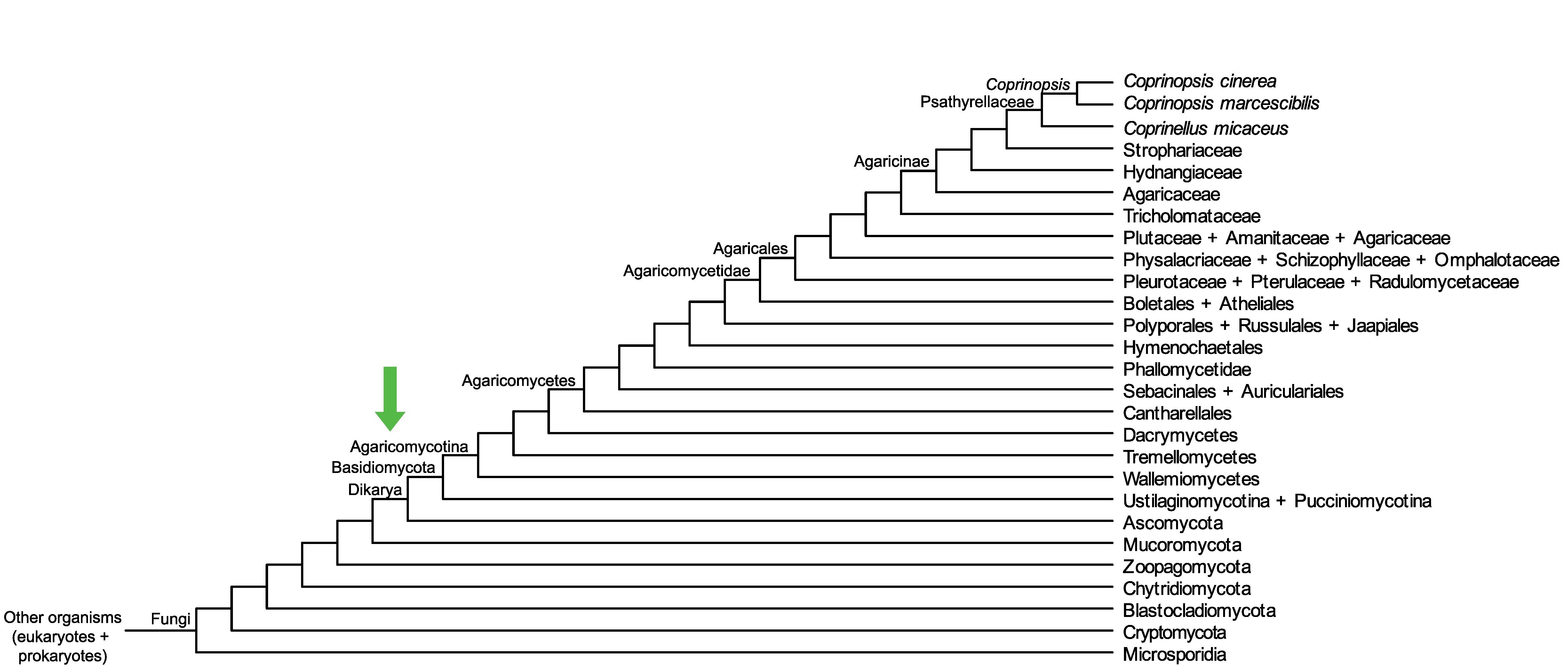
Conservation of CopciAB_470894 across fungi.
Arrow shows the origin of gene family containing CopciAB_470894.
Protein
| Sequence id | CopciAB_470894.T0 |
|---|---|
| Sequence |
>CopciAB_470894.T0 MARSTSRPSTTFTRLLKTVSLIAAASTLSAGVAAYPSKGETLHTLSKREFTLTSPGDLRKSYDYVIVGGGLSGLV LANRLSEDDDRTVLVLEAGLSGDEVKEDVDAPAGAFYNSIVGTDYDWKSMTVNQPGLNNRRIYWPSGKILGGSTA MNAMYIVRPSQPEVDAWAELLESSPDTSSSSSSVERWNWDNLFEAMKKSETFHEPLPEPHEIGGNWPVGEINYGE DGPMQVAFPGVMFSQVAQWLPALEEVGVPSLESPNGGSTLGGFITPSSINPSNWTRSYTRPAYIDSLPPRANLHI LPEALVTRLGFHDKHRSSISGNDRKDQLIANSVEFEVGGDVNRGGEGGRKWVVGVNREVILTAGALGSPKILMHS GVGPRDVLEAAGVDVVHELPGVGQNLQDHMTAGIVFESQMETAGDIHESNSDFARSPEFLSFINDAVAFANITRL FGSDGASALQSELQSTLEALSSGSTPDGLSFSSLLPGASNEVIEGYKATYNLTITTFWSDSVAHVELLMSLISPG TVSIQSALQHPLSRGRVWINSTSPYDPILIDPNYFSHPADRTIMRQGVRLVRELGGVLREMGVVGAEIEPGTEVE SDADLDNWLTNSGATTQYHPMGTCAMLGRELGGVVDEKLRVYGLANVRVADTSVYPYEFAAHLASATFGVAESAA EVVREESFGVPGKARVDERFLGEGGEGSAGGAAVVGRGGLPVVLGVLGVVIANAVLV |
| Length | 732 |
Coding
| Sequence id | CopciAB_470894.T0 |
|---|---|
| Sequence |
>CopciAB_470894.T0 ATGGCCAGGTCAACATCGCGGCCGTCAACGACATTTACGCGTTTACTAAAGACAGTGTCGCTCATCGCGGCTGCA TCGACATTAAGCGCCGGTGTGGCTGCGTATCCCAGCAAGGGTGAAACCCTGCATACCCTCTCAAAACGAGAATTC ACCCTCACTTCCCCTGGCGACCTCCGGAAATCCTACGACTACGTTATTGTAGGCGGCGGATTGTCCGGCCTCGTC CTCGCCAATCGACTAAGCGAGGACGACGATCGTACTGTTCTCGTTCTTGAAGCTGGATTGAGCGGAGATGAGGTG AAGGAGGACGTCGATGCTCCCGCTGGGGCGTTCTACAATTCGATTGTGGGCACGGATTATGATTGGAAATCGATG ACTGTGAACCAGCCTGGGTTGAATAACAGGCGAATTTACTGGCCTAGTGGCAAGATCCTTGGAGGCTCGACGGCT ATGAACGCTATGTACATCGTTCGCCCTTCTCAGCCTGAAGTCGACGCATGGGCAGAGCTGTTGGAATCTTCACCT GACACTTCGTCCTCTTCGTCGTCCGTGGAGAGATGGAACTGGGATAACCTGTTTGAAGCGATGAAGAAGAGCGAA ACGTTCCACGAGCCACTCCCCGAACCTCACGAGATTGGAGGTAACTGGCCAGTCGGAGAGATCAACTATGGCGAA GACGGGCCTATGCAGGTTGCTTTCCCCGGAGTGATGTTCTCCCAAGTCGCACAATGGCTCCCCGCGCTTGAAGAA GTCGGCGTACCTTCCCTCGAATCTCCAAACGGCGGTTCGACACTTGGCGGCTTCATCACCCCGTCCTCGATCAAC CCCTCCAACTGGACGCGCTCATACACACGCCCAGCTTACATCGACTCCCTCCCCCCACGCGCAAACCTCCACATC CTTCCTGAAGCACTCGTCACTCGCCTCGGATTCCACGACAAGCATCGTTCATCGATCTCGGGTAACGATAGGAAG GACCAGCTGATTGCCAACTCGGTCGAGTTTGAGGTGGGAGGTGATGTGAATCGTGGTGGAGAGGGCGGGAGGAAG TGGGTCGTTGGTGTGAATCGGGAGGTTATTTTGACTGCAGGTGCGTTGGGCAGTCCGAAGATTTTGATGCATAGT GGTGTGGGACCTCGGGACGTGCTGGAGGCTGCGGGTGTCGATGTGGTGCACGAGTTGCCCGGTGTTGGCCAGAAC CTTCAGGATCATATGACTGCTGGCATCGTGTTTGAGAGCCAGATGGAGACTGCCGGGGACATCCACGAGTCCAAC TCTGACTTTGCCCGATCCCCCGAGTTCCTCTCGTTCATCAACGACGCGGTAGCCTTCGCGAACATCACCCGCCTC TTCGGATCCGACGGCGCCTCTGCCCTCCAATCCGAACTCCAATCCACACTCGAAGCTCTCTCATCCGGATCGACT CCAGACGGCCTATCCTTCTCCTCCCTCCTCCCCGGCGCCTCCAACGAGGTGATCGAAGGCTACAAGGCAACGTAT AACCTCACGATTACGACCTTCTGGTCCGACTCGGTCGCGCACGTCGAGTTGCTCATGAGCCTCATCTCACCGGGT ACGGTCTCCATCCAGTCTGCGCTGCAGCACCCGCTCAGCCGCGGACGCGTGTGGATCAACTCGACGTCGCCGTAC GACCCTATCCTCATCGACCCCAACTACTTCTCGCACCCTGCTGATCGGACGATTATGCGCCAGGGCGTGAGGTTG GTCAGGGAACTGGGAGGGGTGCTGCGCGAGATGGGGGTCGTAGGAGCCGAGATCGAGCCAGGCACCGAAGTGGAG AGCGACGCTGATCTGGACAATTGGCTAACCAACAGCGGTGCGACGACGCAGTACCACCCGATGGGTACGTGCGCG ATGCTCGGGCGGGAGTTGGGCGGTGTGGTGGATGAGAAGCTGAGGGTGTACGGGTTGGCGAATGTGCGTGTGGCG GATACGAGTGTTTATCCGTACGAGTTTGCGGCGCATTTGGCGAGTGCGACGTTTGGGGTTGCGGAGAGTGCTGCG GAGGTTGTGAGGGAGGAGTCGTTTGGGGTTCCGGGGAAGGCGAGGGTTGATGAGAGGTTTTTGGGTGAGGGTGGG GAGGGGAGTGCGGGTGGTGCGGCTGTGGTGGGGAGGGGTGGTCTCCCGGTGGTGCTTGGTGTGTTGGGTGTTGTG ATTGCGAATGCGGTTTTGGTGTAG |
| Length | 2199 |
Transcript
| Sequence id | CopciAB_470894.T0 |
|---|---|
| Sequence |
>CopciAB_470894.T0 AAACTAGATTCCAGCGATACCCCTACTTATACCAACCAACGACGAACGACACGAACGATACTCGAACTCGATACC ATGGCCAGGTCAACATCGCGGCCGTCAACGACATTTACGCGTTTACTAAAGACAGTGTCGCTCATCGCGGCTGCA TCGACATTAAGCGCCGGTGTGGCTGCGTATCCCAGCAAGGGTGAAACCCTGCATACCCTCTCAAAACGAGAATTC ACCCTCACTTCCCCTGGCGACCTCCGGAAATCCTACGACTACGTTATTGTAGGCGGCGGATTGTCCGGCCTCGTC CTCGCCAATCGACTAAGCGAGGACGACGATCGTACTGTTCTCGTTCTTGAAGCTGGATTGAGCGGAGATGAGGTG AAGGAGGACGTCGATGCTCCCGCTGGGGCGTTCTACAATTCGATTGTGGGCACGGATTATGATTGGAAATCGATG ACTGTGAACCAGCCTGGGTTGAATAACAGGCGAATTTACTGGCCTAGTGGCAAGATCCTTGGAGGCTCGACGGCT ATGAACGCTATGTACATCGTTCGCCCTTCTCAGCCTGAAGTCGACGCATGGGCAGAGCTGTTGGAATCTTCACCT GACACTTCGTCCTCTTCGTCGTCCGTGGAGAGATGGAACTGGGATAACCTGTTTGAAGCGATGAAGAAGAGCGAA ACGTTCCACGAGCCACTCCCCGAACCTCACGAGATTGGAGGTAACTGGCCAGTCGGAGAGATCAACTATGGCGAA GACGGGCCTATGCAGGTTGCTTTCCCCGGAGTGATGTTCTCCCAAGTCGCACAATGGCTCCCCGCGCTTGAAGAA GTCGGCGTACCTTCCCTCGAATCTCCAAACGGCGGTTCGACACTTGGCGGCTTCATCACCCCGTCCTCGATCAAC CCCTCCAACTGGACGCGCTCATACACACGCCCAGCTTACATCGACTCCCTCCCCCCACGCGCAAACCTCCACATC CTTCCTGAAGCACTCGTCACTCGCCTCGGATTCCACGACAAGCATCGTTCATCGATCTCGGGTAACGATAGGAAG GACCAGCTGATTGCCAACTCGGTCGAGTTTGAGGTGGGAGGTGATGTGAATCGTGGTGGAGAGGGCGGGAGGAAG TGGGTCGTTGGTGTGAATCGGGAGGTTATTTTGACTGCAGGTGCGTTGGGCAGTCCGAAGATTTTGATGCATAGT GGTGTGGGACCTCGGGACGTGCTGGAGGCTGCGGGTGTCGATGTGGTGCACGAGTTGCCCGGTGTTGGCCAGAAC CTTCAGGATCATATGACTGCTGGCATCGTGTTTGAGAGCCAGATGGAGACTGCCGGGGACATCCACGAGTCCAAC TCTGACTTTGCCCGATCCCCCGAGTTCCTCTCGTTCATCAACGACGCGGTAGCCTTCGCGAACATCACCCGCCTC TTCGGATCCGACGGCGCCTCTGCCCTCCAATCCGAACTCCAATCCACACTCGAAGCTCTCTCATCCGGATCGACT CCAGACGGCCTATCCTTCTCCTCCCTCCTCCCCGGCGCCTCCAACGAGGTGATCGAAGGCTACAAGGCAACGTAT AACCTCACGATTACGACCTTCTGGTCCGACTCGGTCGCGCACGTCGAGTTGCTCATGAGCCTCATCTCACCGGGT ACGGTCTCCATCCAGTCTGCGCTGCAGCACCCGCTCAGCCGCGGACGCGTGTGGATCAACTCGACGTCGCCGTAC GACCCTATCCTCATCGACCCCAACTACTTCTCGCACCCTGCTGATCGGACGATTATGCGCCAGGGCGTGAGGTTG GTCAGGGAACTGGGAGGGGTGCTGCGCGAGATGGGGGTCGTAGGAGCCGAGATCGAGCCAGGCACCGAAGTGGAG AGCGACGCTGATCTGGACAATTGGCTAACCAACAGCGGTGCGACGACGCAGTACCACCCGATGGGTACGTGCGCG ATGCTCGGGCGGGAGTTGGGCGGTGTGGTGGATGAGAAGCTGAGGGTGTACGGGTTGGCGAATGTGCGTGTGGCG GATACGAGTGTTTATCCGTACGAGTTTGCGGCGCATTTGGCGAGTGCGACGTTTGGGGTTGCGGAGAGTGCTGCG GAGGTTGTGAGGGAGGAGTCGTTTGGGGTTCCGGGGAAGGCGAGGGTTGATGAGAGGTTTTTGGGTGAGGGTGGG GAGGGGAGTGCGGGTGGTGCGGCTGTGGTGGGGAGGGGTGGTCTCCCGGTGGTGCTTGGTGTGTTGGGTGTTGTG ATTGCGAATGCGGTTTTGGTGTAGAATGACGAGTTTGAAGATTCTTGGACACTTTTGGTTCGACGCTTTATTCAC TTGTGATATGGTTTTCTGGTTATGTTCACGACAGACACTGTAACGCTAATGTTTATTGGTTTCTCATTTGGACAC TCTTTAAGTTTTAGTGCTGTTTGTAGTATACCTGTGTAGGGTACTCGCCTCTGAGTAGTCAATTTGTGAAAGATA GCCAAGTCCGG |
| Length | 2486 |
Gene
| Sequence id | CopciAB_470894.T0 |
|---|---|
| Sequence |
>CopciAB_470894.T0 AAACTAGATTCCAGCGATACCCCTACTTATACCAACCAACGACGAACGACACGAACGATACTCGAACTCGATACC ATGGCCAGGTCAACATCGCGGCCGTCAACGACATTTACGCGTTTACTAAAGACAGTGTCGCTCATCGCGGCTGCA TCGACATTAAGCGCCGGTGTGGCTGCGTATCCCAGCAAGGGTGAAACGTGCGTAACGCGTGTCTTCTCCTCCTCG AATACATTCTAACCCCCTTCACCGCAGCCTGCATACCCTCTCAAAACGAGAATTCACCCTCACTTCCCCTGGCGA CCTCCGGAAATCCTACGACTACGTTATTGTAGGCGGCGGATTGTCCGGCCTCGTCCTCGCCAATCGACTAAGCGA GGACGACGATCGTACTGTTCTCGTTCTTGAAGCTGGATTGAGCGGAGATGAGGTGAAGGAGGACGTCGATGCTCC CGCTGGGGCGTTCTACAATTCGATTGTGGGCACGGATTATGATTGGAAATCGATGACTGTGAACCAGCCTGGGTT GAATAACAGGCGAATTTACTGGCCTAGTGGCAAGGTGCGTCGTTTTCTGATACTTTTCGTGCTGGTACTGAGCGT CTTTGGCTTTAGATCCTTGGAGGCTCGACGGCTATGAACGCTATGTACATCGTTCGCCCTTCTCAGCCTGAAGTC GACGCATGGGCAGAGCTGTTGGAATCTTCACCTGACACTTCGTCCTCTTCGTCGTCCGTGGAGAGATGGAACTGG GATAACCTGTTTGAAGCGATGAAGAAGAGCGAAACGTTCCACGAGCCACTCCCCGAACCTCACGAGATTGGAGGT AACTGGCCAGTCGGAGAGATCAACTATGGCGAAGACGGGCCTATGCAGGTTGCTTTCCCCGGAGTGTACGTCCCA AACTTCTCTCCTTGTTCATCTGAAATTAACCAATAACCTACTACAGGATGTTCTCCCAAGTCGCACAATGGCTCC CCGCGCTTGAAGAAGTCGGCGTACCTTCCCTCGAATCTCCAAACGGCGGTTCGACACTTGGCGGCTTCATCACCC CGTCCTCGATCAACCCCTCCAACTGGACGCGCTCATACACACGCCCAGCTTACATCGACTCCCTCCCCCCACGCG CAAACCTCCACATCCTTCCTGAAGCACTCGTCACTCGCCTCGGATTCCACGACAAGCATCGTTCATCGATCTCGG GTAACGATAGGAAGGACCAGCTGATTGCCAACTCGGTCGAGTTTGAGGTGGGAGGTGATGTGAATCGTGGTGGAG AGGGCGGGAGGAAGTGGGTCGTTGGTGTGAATCGGGAGGTTATTTTGACTGCAGGTGCGTTGGGCAGTCCGAAGA TTTTGATGCATAGTGGTGTGGGACCTCGGGACGTGCTGGAGGCTGCGGGTGTCGATGTGGTGCACGAGTTGCCCG GTGTTGGCCAGAACCTTCAGGATCATATGGTGAGTTAATTATCGATATCTGAAAACTCGATTCTAATCACATTTC CAGACTGCTGGCATCGTGTTTGAGAGCCAGATGGAGACTGCCGGGGACATCCACGAGTCCAACTCTGACTTTGCC CGATCCCCCGAGTTCCTCTCGTTCATCAACGACGCGGTAGCCTTCGCGAACATCACCCGCCTCTTCGGATCCGAC GGCGCCTCTGCCCTCCAATCCGAACTCCAATCCACACTCGAAGCTCTCTCATCCGGATCGACTCCAGACGGCCTA TCCTTCTCCTCCCTCCTCCCCGGCGCCTCCAACGAGGTGATCGAAGGCTACAAGGCAACGTATAACCTCACGATT ACGACCTTCTGGTCCGACTCGGTCGCGCACGTCGAGTTGCTCATGAGCCTCATCTCACCGGGTACGGTCTCCATC CAGTCTGCGCTGCAGCACCCGCTCAGCCGCGGACGCGTGTGGATCAACTCGACGTCGCCGTACGACCCTATCCTC ATCGACCCCAACTACTTCTCGCACCCTGCTGATCGGACGATTATGCGCCAGGGCGTGAGGTTGGTCAGGGAACTG GGAGGGGTGCTGCGCGAGATGGGGGTCGTAGGAGCCGAGATCGAGCCAGGCACCGAAGTGGAGAGCGACGCTGAT CTGGACAATTGGCTAACCAACAGCGGTGCGACGACGCAGTACCACCCGATGGGTACGTGCGCGATGCTCGGGCGG GAGTTGGGCGGTGTGGTGGATGAGAAGCTGAGGGTGTACGGGTTGGCGAATGTGCGTGTGGCGGATACGAGTGTT TATCCGTACGAGTTTGCGGCGCATTTGGCGAGTGCGACGTTTGGGGTTGCGGAGAGTGCTGCGGAGGTTGTGAGG GAGGAGTCGTTTGGGGTTCCGGGGAAGGCGAGGGTTGATGAGAGGTTTTTGGGTGAGGGTGGGGAGGGGAGTGCG GGTGGTGCGGCTGTGGTGGGGAGGGGTGGTCTCCCGGTGGTGCTTGGTGTGTTGGGTGTTGTGATTGCGAATGCG GTTTTGGTGTAGAATGACGAGTTTGAAGATTCTTGGACACTTTTGGTTCGACGCTTTATTCACTTGTGATATGGT TTTCTGGTTATGTTCACGACAGACACTGTAACGCTAATGTTTATTGGTTTCTCATTTGGACACTCTTTAAGTTTT AGTGCTGTTTGTAGTATACCTGTGTAGGGTACTCGCCTCTGAGTAGTCAATTTGTGAAAGATAGCCAAGTCCGG |
| Length | 2699 |