CopciAB_471204
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_471204 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 471204 |
| Uniprot id | Functional description | Kinase-like | |
| Location | scaffold_3:1613103..1616053 | Strand | - |
| Gene length (nt) | 2951 | Transcript length (nt) | 2283 |
| CDS length (nt) | 1506 | Protein length (aa) | 501 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN7537_ppk33 |
| Neurospora crassa | stk-49 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7265535 | 80.6 | 2.199E-233 | 721 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26329 | 76.6 | 2.937E-222 | 689 |
| Schizophyllum commune H4-8 | Schco3_2503244 | 77.5 | 1.394E-221 | 687 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1388760 | 74.8 | 1.029E-214 | 667 |
| Flammulina velutipes | Flave_chr11AA00551 | 72.5 | 2.06E-205 | 640 |
| Grifola frondosa | Grifr_OBZ72473 | 70.6 | 2.046E-205 | 640 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_58379 | 70.2 | 1.098E-203 | 635 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_191171 | 69.2 | 2.771E-199 | 622 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114943 | 69.2 | 2.887E-199 | 622 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1085758 | 77.5 | 5.845E-192 | 601 |
| Pleurotus ostreatus PC9 | PleosPC9_1_105518 | 77.5 | 5.722E-192 | 601 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_17224 | 67.1 | 1.634E-190 | 597 |
| Lentinula edodes NBRC 111202 | Lenedo1_1163469 | 66.9 | 3.65E-190 | 596 |
| Lentinula edodes B17 | Lened_B_1_1_18565 | 79 | 7.03E-130 | 421 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_471204.T0 |
| Description | Kinase-like |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 22 | 292 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR017441 | Protein kinase, ATP binding site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR000961 | AGC-kinase, C-terminal |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K08793 |
EggNOG
| COG category | Description |
|---|---|
| T | Kinase-like |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
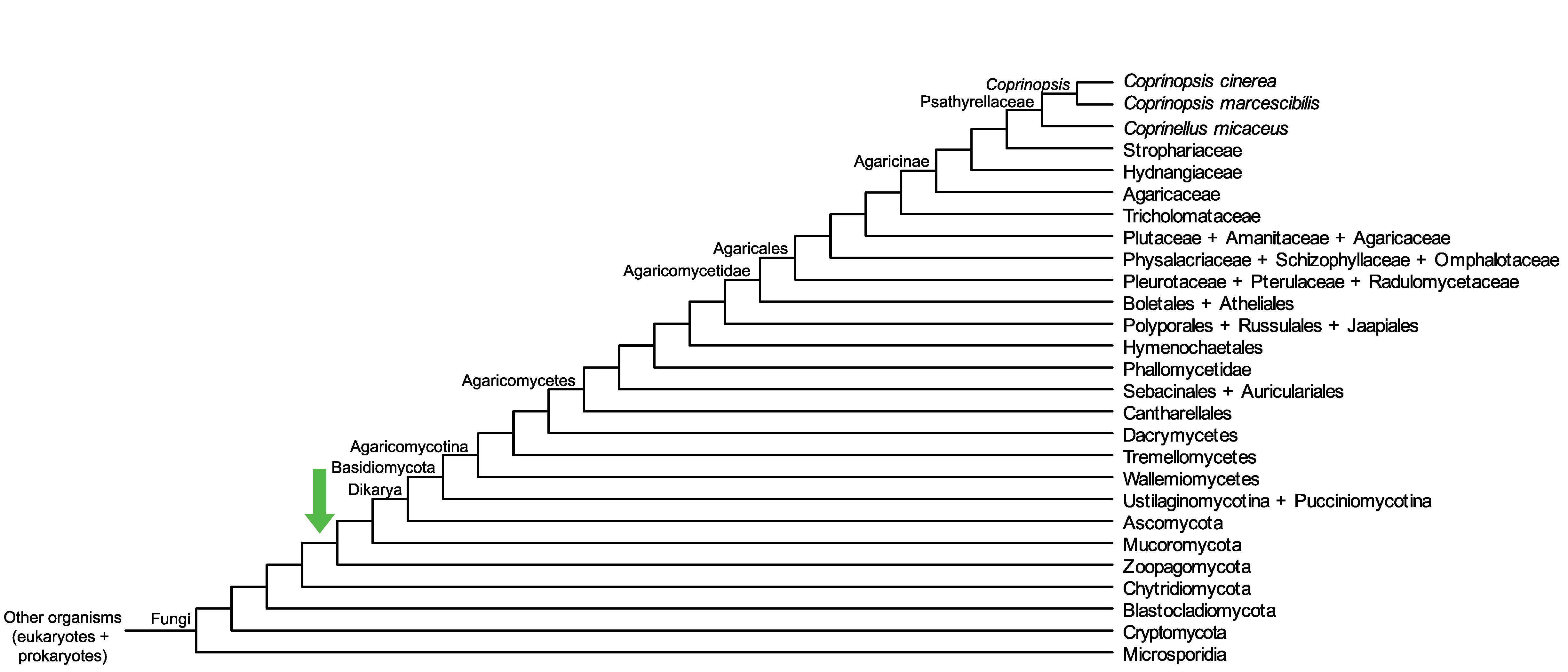
Conservation of CopciAB_471204 across fungi.
Arrow shows the origin of gene family containing CopciAB_471204.
Protein
| Sequence id | CopciAB_471204.T0 |
|---|---|
| Sequence |
>CopciAB_471204.T0 MGCCFSEPVDFDGEVNLYHFDLHRAVGKGAFGKVRVVEHKRSKKLYALKYIDKTRCIKQKAVANIIQERRLLEEI DHPFVVNLRYAFQDDENCFFVLDLMLGGDLRFHLERKGHIPENVVRFWVAELSSAVDYLHRQRIIHRDIKPDNIL LDSEGHAHLTDFNVAIHYSSSRLHSSVAGSMAYMAPQVVSRKGYSWQIDWWSLGVTAYELIYHRRPFDGRNAEKM TQSIMKDTLKFPGRESGEDEKEREKESWCSDDGVSALRAFIERDPHQRLGCKPDARGLEDVKRHPWFRKIDWDAL ESKTIQPPFVPDMKQANFDVSHELDEFLMVEKPLTHSKRKANPDLEKMKPELRQLEEQFTVYDFTSSKRLSYYPH NQPIVALTGTESGDTDRTLAGTIIATQTMVERSQAGSPIQGLESQGGYDGGYQRHQMSIDQQRGYTGSVGVGSTN GHARSGSTRTYEGSQHQQRRQSIQYSRPQRPQHAHIQHQQYQHPSQPRPSS |
| Length | 501 |
Coding
| Sequence id | CopciAB_471204.T0 |
|---|---|
| Sequence |
>CopciAB_471204.T0 ATGGGATGCTGTTTCTCTGAGCCAGTAGATTTCGACGGCGAGGTCAACCTCTATCACTTTGACCTCCACCGCGCT GTGGGGAAGGGCGCTTTTGGAAAGGTCCGCGTTGTTGAACACAAGCGATCCAAGAAGCTCTATGCCCTCAAGTAC ATCGACAAGACACGCTGCATAAAACAGAAGGCTGTCGCGAACATCATCCAAGAACGGAGATTGCTCGAAGAAATC GACCACCCTTTCGTCGTCAACCTGCGCTATGCATTCCAGGACGACGAGAACTGCTTTTTCGTCCTCGACTTGATG TTGGGTGGCGACCTCCGATTCCACTTGGAACGCAAAGGCCACATCCCGGAGAACGTCGTGCGATTCTGGGTAGCT GAGCTCTCCAGCGCGGTTGACTACCTGCACCGACAGCGGATAATCCATCGTGATATCAAGCCCGACAACATCCTC CTCGACTCGGAAGGTCATGCCCACCTCACCGACTTCAACGTCGCAATCCACTACTCCTCGTCACGTCTACATTCC TCCGTGGCTGGCTCCATGGCCTACATGGCACCCCAGGTGGTGTCACGGAAGGGATACAGTTGGCAGATTGATTGG TGGAGTTTGGGAGTCACAGCCTACGAGCTGATCTACCACAGGCGACCGTTTGATGGTCGGAATGCAGAGAAGATG ACACAGAGTATCATGAAGGATACATTGAAGTTCCCTGGAAGGGAAAGCGGAGAGGACGAGAAAGAGAGGGAAAAG GAGTCGTGGTGCAGTGACGATGGGGTTAGCGCTTTGAGAGCGTTCATCGAACGAGACCCTCATCAGCGCCTAGGT TGCAAACCGGACGCACGCGGTTTGGAAGATGTGAAACGACATCCGTGGTTTAGAAAAATCGACTGGGACGCGCTC GAGAGCAAGACGATCCAGCCACCGTTTGTACCTGACATGAAGCAAGCCAACTTTGACGTCTCACACGAACTGGAC GAGTTCCTCATGGTCGAAAAGCCGCTTACCCACTCCAAACGAAAGGCCAACCCCGATCTCGAGAAAATGAAGCCT GAACTGCGTCAACTGGAGGAGCAATTCACCGTGTATGACTTTACATCTTCCAAACGCCTGTCTTATTACCCCCAC AACCAGCCCATAGTTGCACTCACAGGGACCGAGTCTGGTGACACCGACCGGACGCTGGCGGGTACGATTATTGCA ACGCAGACTATGGTTGAGCGGAGTCAGGCAGGATCGCCTATCCAGGGCTTGGAGAGCCAAGGAGGGTATGACGGA GGTTACCAGAGGCATCAAATGTCGATTGATCAGCAGAGAGGGTACACTGGGAGTGTAGGTGTGGGTAGTACGAAT GGGCATGCAAGGAGCGGGAGTACAAGGACATACGAAGGAAGTCAACACCAGCAAAGAAGACAGTCGATCCAGTAT TCAAGGCCGCAGAGGCCGCAACATGCGCATATTCAACACCAACAATATCAGCATCCGTCACAACCACGGCCGTCC TCGTAA |
| Length | 1506 |
Transcript
| Sequence id | CopciAB_471204.T0 |
|---|---|
| Sequence |
>CopciAB_471204.T0 ACACCTTACAGTCTCTTTTTTCTCTTCTCTCTTTTCCTCTTATCTTTACTTCTCCTAATTCACGAAGCATGGGAT GCTGTTTCTCTGAGCCAGTAGATTTCGACGGCGAGGTCAACCTCTATCACTTTGACCTCCACCGCGCTGTGGGGA AGGGCGCTTTTGGAAAGGTCCGCGTTGTTGAACACAAGCGATCCAAGAAGCTCTATGCCCTCAAGTACATCGACA AGACACGCTGCATAAAACAGAAGGCTGTCGCGAACATCATCCAAGAACGGAGATTGCTCGAAGAAATCGACCACC CTTTCGTCGTCAACCTGCGCTATGCATTCCAGGACGACGAGAACTGCTTTTTCGTCCTCGACTTGATGTTGGGTG GCGACCTCCGATTCCACTTGGAACGCAAAGGCCACATCCCGGAGAACGTCGTGCGATTCTGGGTAGCTGAGCTCT CCAGCGCGGTTGACTACCTGCACCGACAGCGGATAATCCATCGTGATATCAAGCCCGACAACATCCTCCTCGACT CGGAAGGTCATGCCCACCTCACCGACTTCAACGTCGCAATCCACTACTCCTCGTCACGTCTACATTCCTCCGTGG CTGGCTCCATGGCCTACATGGCACCCCAGGTGGTGTCACGGAAGGGATACAGTTGGCAGATTGATTGGTGGAGTT TGGGAGTCACAGCCTACGAGCTGATCTACCACAGGCGACCGTTTGATGGTCGGAATGCAGAGAAGATGACACAGA GTATCATGAAGGATACATTGAAGTTCCCTGGAAGGGAAAGCGGAGAGGACGAGAAAGAGAGGGAAAAGGAGTCGT GGTGCAGTGACGATGGGGTTAGCGCTTTGAGAGCGTTCATCGAACGAGACCCTCATCAGCGCCTAGGTTGCAAAC CGGACGCACGCGGTTTGGAAGATGTGAAACGACATCCGTGGTTTAGAAAAATCGACTGGGACGCGCTCGAGAGCA AGACGATCCAGCCACCGTTTGTACCTGACATGAAGCAAGCCAACTTTGACGTCTCACACGAACTGGACGAGTTCC TCATGGTCGAAAAGCCGCTTACCCACTCCAAACGAAAGGCCAACCCCGATCTCGAGAAAATGAAGCCTGAACTGC GTCAACTGGAGGAGCAATTCACCGTGTATGACTTTACATCTTCCAAACGCCTGTCTTATTACCCCCACAACCAGC CCATAGTTGCACTCACAGGGACCGAGTCTGGTGACACCGACCGGACGCTGGCGGGTACGATTATTGCAACGCAGA CTATGGTTGAGCGGAGTCAGGCAGGATCGCCTATCCAGGGCTTGGAGAGCCAAGGAGGGTATGACGGAGGTTACC AGAGGCATCAAATGTCGATTGATCAGCAGAGAGGGTACACTGGGAGTGTAGGTGTGGGTAGTACGAATGGGCATG CAAGGAGCGGGAGTACAAGGACATACGAAGGAAGTCAACACCAGCAAAGAAGACAGTCGATCCAGTATTCAAGGC CGCAGAGGCCGCAACATGCGCATATTCAACACCAACAATATCAGCATCCGTCACAACCACGGCCGTCCTCGTAAA CGCATTATATCTCAAGTCCCTGAGAACTTGTTATGATCTGGCTATGATATTCACTCACATTTGCGGTTATGGCTT TGGGTTGTGTTATGCGCTCCGGGCGGACGTACACTCGACTATGGAGTACTTTCCTATATCCATGCATCCAGCTTT CTTTACCATACATTTTGTGCCCTCTTAATACCTTTTTCTACTCTACCCGTCTACTCATATCATGCTTTCCCCGGC TTCTACATCATATTTGACCCTGCACACACCCTCATGTTTACATTCGACATCTTCGCATCATCTGCATTCTTCATA CTTTTCGATTTGTACTGCTTCAATCCTACTTGCACCGCACCCATCCACACGCATACTCACTTCACTATTCCACCT AGTATACGAATTACGACGACTCCCCTTTTCTTTGATTCTTGTCTACGATCATGTCATACCCCCCTCCTTTGCTAT CCTTGCTATCGCAATTCCGCTCACCCGCGTTCCGCAGGCATGTACATACACATAAACATCTGTACCATTATTCGT CGTTGTCGTGGTTCTACGTATTACTTTTCCTTTCCCCTCCTCCCCTGTTTTTCTACTCTTCGCTTCGCCTCGCAT GAGACGCTGGATTTGCAGTTCACAACTTCTATCATTTACATAGCTTATTTCCCTCCTTTACGTTGTGTAATTTTA GGTAGATGGTATATGGAATTCTTTGGAAATCGG |
| Length | 2283 |
Gene
| Sequence id | CopciAB_471204.T0 |
|---|---|
| Sequence |
>CopciAB_471204.T0 ACACCTTACAGTCTCTTTTTTCTCTTCTCTCTTTTCCTCTTATCTTTACTTCTCCTAATTCACGAAGCATGGGAT GCTGTTTCTCTGAGCCAGTAGATTTCGACGGCGAGGTCAACCTCTATCACTTTGACCTCCACCGCGCTGTGGGGA AGGGCGCTTTTGGAAAGGTCCGTTCTGCTTCGATTTGCCAGAACCGACATGACCACTAATCTCGCTGTCCAGGTC CGCGTTGTTGAACACAAGCGATCCAAGAAGCTCTATGCCCTCAAGTACATCGACAAGACACGCTGCATAAAACAG AAGGCTGTCGCGAACATCATCCAAGAACGGAGATTGCTCGAAGAAGTACGCACATTCCAGCATGTTCTTCCCACT TCTTGTCCCTGACGATCCTACCCTGCCCACATTCAGATCGACCACCCTTTCGTCGTCAACCTGCGCTATGCATTC CAGGACGACGAGAACTGCTTTTTCGTCCTCGACTTGATGTTGGGTGGCGACCTCCGATGTTCGTCCGCCGTTTGC TTGCATTTTCTTACATTCCACTCAACGTCCAACCAACAGTCCACTTGGAACGCAAAGGCCACATCCCGGAGAACG TCGTGCGATTCTGGGTAGCTGAGCTCTCCAGCGCGGTTGACTACCTGCACCGACAGCGGATAATCCATCGGTACG CGCCATCTCACCATTCCACCTCTTCACATGTTCGCTAACGACCGCCTTGATCCTAACAATCATAATCATATGGCT CCCCTGTCCCCCTGACTCTATCCCCGGCCTCCTACACCGCCCCTCGCCCCTCTGATTCACATTACACAGTGATAT CAAGCCCGACAACATCCTCCTCGACTCGGAAGGTCATGCCCACCTCACCGACTTCAACGTCGCAATCCACTACTC CTCGTCACGTCTACATTCCTCCGTGGCTGGCTCCATGGCCTACATGGCACCCCAGGTGGTGTCACGGAAGGGATA CAGTTGGCAGATTGATTGGTGGAGTTTGGGAGTCACAGCCTACGAGCTGATCTACCACAGGCGACCGTTTGATGG TCGGAATGCAGAGAAGATGACACAGAGTATCATGAAGGATACATTGAAGTTCCCTGGAAGGGAAAGCGGAGAGGA CGAGAAAGAGAGGGAAAAGGAGTCGTGGTGCAGTGACGATGGGGTTAGCGCTTTGAGAGCGGTGAGTGACATCGG ATGATCTCCTTTGGCGAATCTAATTGGCTATCGGTTTACAGTTCATCGAACGAGACCCTCATCAGCGCCTAGGTT GCAAACCGGACGCACGCGGTTTGGAAGATGTGAAACGACATCCGTGGTTTAGAAAAATCGACTGGGACGCGCTCG AGAGCAAGACGATCCAGCCACCGTTTGTACCTGACGTGCGTCTCGTTCCTGTCTTCTTTGGCCACCATCTTTTCC CCCATACATTTGGCGCCCACATACCCACTGTCGGAGTTGCCACACCCGTGCATCAATCATTCTTCCCTGCTTCCA CTTTCCCATATAGACGTTTATCGAGAGCGATCGTACTGACGCGAAGCTCGCGTATGAACAGATGAAGCAAGCCAA CTTTGACGTCTCACACGAACTGGACGAGTTCCTCATGGTCGAAAAGCCGCTTACCCACTCCAAACGAAAGGCCAA CCCCGATCTCGAGAAAATGAAGCCTGAACTGCGTCAACTGGAGGAGCAGTACGTCCTGGTCTTCATTTCTTCTAC TAACACTAATGCACTACCTCAGATTCACCGTGTATGACTTTACATCTTCCAAACGCCTGTCTTATTACCCCCACA ACCAGCCCATAGTTGCACTCACAGGGACCGAGTCTGGTGACACCGACCGGACGCTGGCGGGTACGATTATTGCAA CGCAGACTATGGTTGAGCGGAGTCAGGCAGGATCGCCTATCCAGGGCTTGGAGAGCCAAGGAGGGTATGACGGAG GTTACCAGAGGCATCAAATGTCGATTGATCAGCAGAGAGGGTACACTGGGAGTGTAGGTGTGGGTAGTACGAATG GGCATGCAAGGAGCGGGAGTACAAGGACATACGAAGGAAGTCAACACCAGCAAAGAAGACAGTCGATCCAGTATT CAAGGCCGCAGAGGCCGCAACATGCGCATATTCAACACCAACAATATCAGCATCCGTCACAACCACGGCCGTCCT CGTAAACGCATTATATCTCAAGTCCCTGAGAACTTGTTATGATCTGGCTATGATATTCACTCACATTTGCGGTTA TGGCTTTGGGTGAGTATCTTGTTCCTTTTGCGCCTGGGGATGGGAAGCTGATGTGTTCCTGGGCGCTATAGGTTG TGTTATGCGCTCCGGGCGGACGTACACTCGACTATGGAGTACTTTCCTATATCCATGCATCCAGCTTTCTTTACC ATACATTTTGTGCCCTCTTAATACCTTTTTCTACTCTACCCGTCTACTCATATCATGCTTTCCCCGGCTTCTACA TCATATTTGACCCTGCACACACCCTCATGTTTACATTCGACATCTTCGCATCATCTGCATTCTTCATACTTTTCG ATTTGTACTGCTTCAATCCTACTTGCACCGCACCCATCCACACGCATACTCACTTCACTATTCCACCTAGTATAC GAATTACGACGACTCCCCTTTTCTTTGATTCTTGTCTACGATCATGTCATACCCCCCTCCTTTGCTATCCTTGCT ATCGCAATTCCGCTCACCCGCGTTCCGCAGGCATGTACATACACATAAACATCTGTACCATTATTCGTCGTTGTC GTGGTTCTACGTATTACTTTTCCTTTCCCCTCCTCCCCTGTTTTTCTACTCTTCGCTTCGCCTCGCATGAGACGC TGGATTTGCAGTTCACAACTTCTATCATTTACATAGCTTATTTCCCTCCTTTACGTTGTGTAATTTTAGGTAGAT GGTATATGGAATTCTTTGGAAATCGG |
| Length | 2951 |