CopciAB_471344
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_471344 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 471344 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_4:2102111..2106367 | Strand | + |
| Gene length (nt) | 4257 | Transcript length (nt) | 3952 |
| CDS length (nt) | 3156 | Protein length (aa) | 1051 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7259222 | 60 | 0 | 1193 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_117856 | 59.7 | 0 | 1139 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_37217 | 59.7 | 0 | 1138 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB24910 | 59.8 | 0 | 1136 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_6963 | 57 | 0 | 1070 |
| Pleurotus ostreatus PC9 | PleosPC9_1_133080 | 55.4 | 0 | 1047 |
| Lentinula edodes NBRC 111202 | Lenedo1_1045551 | 58.6 | 0 | 1030 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1445012 | 54.9 | 0 | 1030 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1093414 | 59.4 | 0 | 1016 |
| Flammulina velutipes | Flave_chr07AA00366 | 54.1 | 5.838E-292 | 951 |
| Grifola frondosa | Grifr_OBZ67088 | 51.4 | 1.836E-275 | 871 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_116244 | 54 | 2.84E-245 | 783 |
| Auricularia subglabra | Aurde3_1_1008339 | 43 | 7.079E-198 | 645 |
| Schizophyllum commune H4-8 | Schco3_1039150 | 37.4 | 1.671E-133 | 454 |
| Lentinula edodes B17 | Lened_B_1_1_15076 | 57.2 | 1.92E-83 | 301 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_471344.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 220 | 445 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
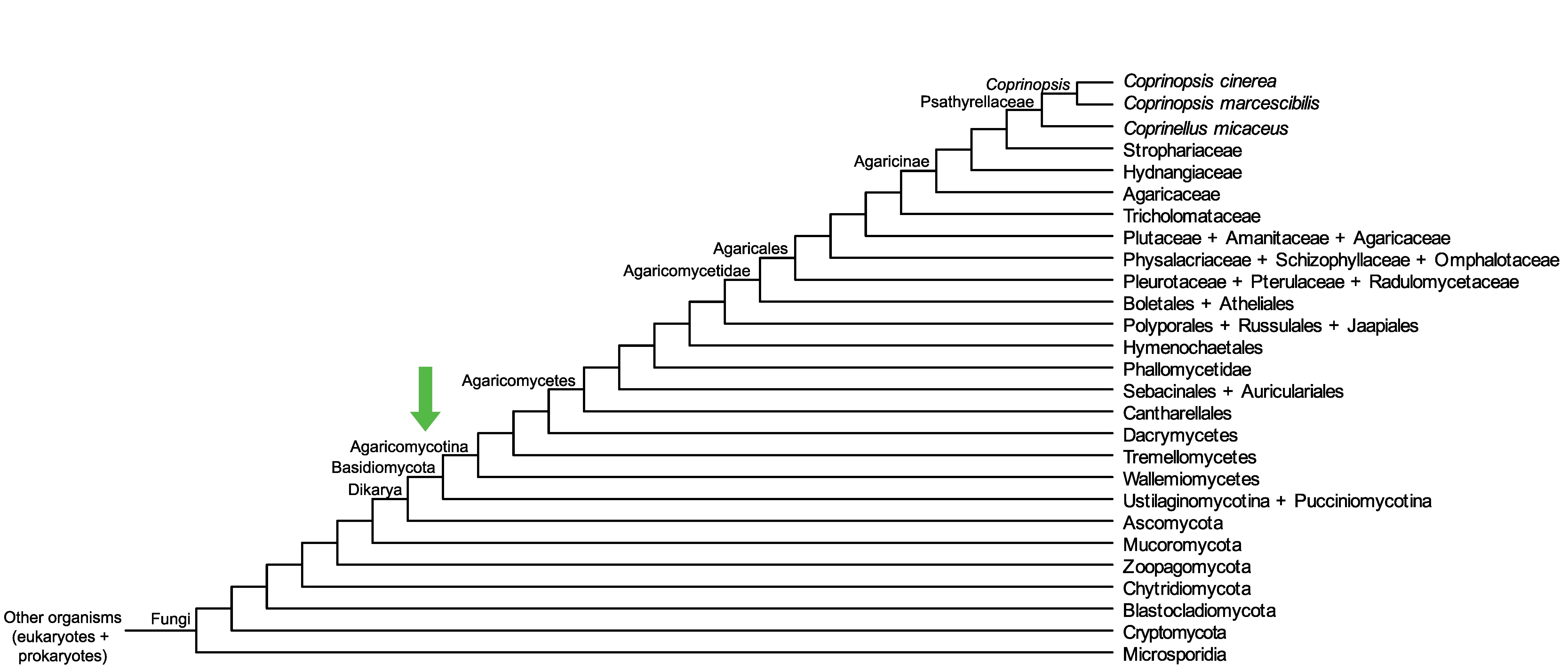
Conservation of CopciAB_471344 across fungi.
Arrow shows the origin of gene family containing CopciAB_471344.
Protein
| Sequence id | CopciAB_471344.T0 |
|---|---|
| Sequence |
>CopciAB_471344.T0 MLQYPVQVPSLSLSSSAKLASKPSKDNIPYLLSNDPPRQPQPLSSLRFQKDPYAARPVGPNRLPAPIVPLSISTN LVPSPSASSPSTASAKHPRSPYISSRQLSASPVPFRQQPSGPAPPSALSQCASSGSYPIEDVLTPGDIVGEGLVL QNEVLRLVGSTNESDAAAPAKEFEVVRVLGTGSYAVVYLVREILSRPPPSEDGHMTTIGSMDMDGTPQLEYGREY AIKLLSKANLDEEALAAQMTEVTIHQSLPLHPNIVTLHRTLETPSFLLLLLEYVPGEDLFYFLEQARDHYEPQPS DFVMYNSRTPPTPSLLSNLHPSQLLSRTRLRLIASMFSQMCDAVAACHSTQVFHRDIKPENFIVTDGWVMLANGR QERKVIVKLTDFGLSTTDIDSSDVDCGSAPYMSYECRNNIAPTYRPRDADVWSLGIVLINMLYHFNPWTDTAEGA CSSFDLYRRDPVNFFTNRFPGMTRGVAEFLASRVFCILEGDESEGRRISAADFGVWVKDNLVTMLAGHGMQRTLS TSSTQGHPITSIPVSHRPMSRNPSNATAARGTPAMPTRSLSRAPSLGPAYEREEKSELSTVIDQDNEDDVEVPEP SEPILGVEEAGSRSASTSKRRKRGARKGKGAQASSPTSPQDDTIVNLAAASECLARELSKQSKVSLPSSPKSTST SSRKRQPLEPVSMYAMPTPLLNSSTPRTATKASTFVTCTPVPPSAPTKKPSRWKLSFGKANASTLANAVERPATE EAASPASAEPSQPPPPSTTVNNVTNLIMGLDAPAARPIPPAAGVFDDGASTWARGRRPRDASPVRKYGGRSPRRG LQSPSQPSLADSWVDPPSRGPSRAISPSSTRSGRPIRSSASSMASSNWRSSMSTTSSAGTSTSAFTHYSNGSTRS ISTTATSVSATSWRTSVKQPSISSNSSGYSASGGYTSVPKNIKLMNGVPWELDQLPRGQHPDPSFGNPPPRKQRT RKHKDIKLDTITERPSNSAGLKSPELRKDAMTSTSDLPAVVNAARDEGDNVGSGGVKKVHKGQINALAKMLSALR R |
| Length | 1051 |
Coding
| Sequence id | CopciAB_471344.T0 |
|---|---|
| Sequence |
>CopciAB_471344.T0 ATGTTGCAGTACCCGGTTCAGGTTCCCTCGCTGTCCCTCTCCTCATCTGCAAAGCTCGCCTCTAAACCCTCTAAA GATAACATTCCTTATTTGCTCAGTAATGACCCCCCTCGCCAGCCACAGCCATTGAGCTCCCTCCGCTTCCAGAAG GACCCGTATGCTGCCAGGCCAGTTGGTCCAAATCGTTTGCCAGCACCGATCGTGCCACTCTCGATTTCGACCAAT CTGGTCCCGTCACCGTCAGCTTCGTCCCCGTCAACCGCCTCGGCCAAGCACCCACGTAGCCCTTACATCTCCTCC CGTCAGCTTTCTGCAAGCCCCGTCCCGTTCCGCCAGCAACCCAGCGGCCCAGCACCCCCTTCGGCTCTATCCCAA TGCGCCTCTTCAGGCTCGTATCCCATTGAAGACGTGTTAACACCTGGGGATATTGTCGGCGAGGGCCTTGTTCTC CAGAATGAGGTTCTCCGGTTGGTTGGGTCCACGAACGAGTCAGATGCAGCTGCACCAGCCAAGGAGTTTGAAGTC GTGCGTGTCCTCGGCACTGGCAGCTACGCTGTTGTCTACCTGGTTCGAGAAATTCTTTCCCGACCCCCTCCTTCT GAAGACGGCCACATGACAACCATCGGCTCCATGGATATGGACGGCACCCCTCAGCTAGAGTATGGGCGTGAATAC GCTATCAAATTGCTGTCCAAGGCCAACCTTGACGAGGAGGCTCTGGCCGCCCAAATGACTGAGGTTACGATCCAT CAATCTCTTCCCCTCCACCCAAATATTGTAACGCTACACCGCACACTTGAGACCCCTTCTTTCTTACTTCTTCTC CTGGAATACGTTCCGGGCGAGGACCTTTTCTATTTTTTGGAACAAGCCCGTGACCATTATGAACCCCAGCCTTCT GATTTTGTCATGTACAACTCTCGCACTCCCCCAACTCCCAGTTTACTCTCCAATCTCCACCCTTCGCAACTGCTC TCGCGCACTCGCCTTCGCCTCATCGCTTCGATGTTCTCCCAGATGTGTGACGCTGTTGCCGCCTGCCACTCCACC CAAGTCTTCCACCGCGATATCAAGCCTGAAAACTTTATCGTTACAGATGGTTGGGTGATGCTGGCGAACGGACGT CAAGAACGAAAGGTCATTGTCAAGTTGACGGACTTCGGTTTGTCAACTACCGACATCGACTCTTCGGACGTCGAT TGTGGAAGCGCGCCCTACATGAGCTACGAATGCAGGAACAACATTGCTCCCACCTATCGACCTCGCGATGCTGAT GTTTGGTCTCTTGGCATCGTGCTAATCAACATGTTGTATCACTTTAACCCATGGACCGACACGGCCGAAGGTGCT TGCTCGTCGTTCGACCTTTACCGTCGGGATCCCGTCAATTTCTTCACCAATCGTTTCCCCGGAATGACTCGAGGT GTTGCTGAGTTCTTGGCTTCAAGAGTCTTTTGCATCCTCGAGGGTGATGAGAGCGAGGGCCGACGCATCTCTGCG GCCGATTTCGGTGTTTGGGTCAAGGATAACTTGGTGACGATGTTGGCTGGCCATGGGATGCAGCGCACTTTGTCC ACCAGTTCGACGCAAGGTCATCCTATTACCTCCATCCCCGTGTCTCATCGTCCCATGTCCCGTAATCCCTCCAAC GCGACCGCCGCTCGTGGTACTCCTGCCATGCCCACTAGATCGCTTTCCCGGGCACCATCCCTCGGCCCCGCCTAC GAACGCGAGGAGAAATCAGAGCTGTCCACTGTCATCGATCAAGATAACGAAGATGATGTCGAGGTGCCCGAGCCT TCTGAACCTATCCTCGGCGTCGAAGAAGCTGGCTCTCGATCTGCCTCCACTTCCAAGCGTCGTAAGCGCGGAGCT CGCAAAGGCAAAGGTGCCCAAGCTTCGTCGCCTACATCCCCCCAAGACGATACAATTGTCAACCTGGCCGCGGCT TCTGAGTGTTTGGCTCGGGAGCTGAGCAAGCAGTCGAAGGTCTCCCTTCCCTCTTCACCCAAGAGCACTTCCACC TCCTCGCGCAAGCGACAGCCCCTCGAACCCGTATCCATGTACGCCATGCCCACCCCTCTGCTTAATTCTTCTACC CCTCGAACTGCAACGAAAGCTTCAACATTTGTCACTTGCACTCCTGTACCACCCTCGGCTCCAACGAAGAAGCCA TCCCGGTGGAAGCTCAGCTTTGGTAAGGCCAACGCATCAACCTTGGCAAACGCCGTCGAGCGGCCTGCGACTGAA GAAGCTGCATCACCAGCTTCTGCTGAACCATCTCAACCACCACCTCCGTCTACCACTGTCAACAACGTCACGAAT CTTATCATGGGCTTGGATGCTCCTGCTGCCCGTCCTATTCCTCCTGCTGCAGGTGTATTTGACGATGGTGCATCT ACTTGGGCACGTGGCCGTCGACCAAGAGACGCCTCCCCTGTGCGCAAGTATGGTGGACGGTCTCCCCGACGAGGA CTTCAATCTCCTTCCCAGCCCAGCTTGGCTGACAGTTGGGTTGACCCTCCTTCGCGCGGTCCTTCACGTGCCATC TCTCCTAGCAGTACGCGAAGCGGGCGTCCGATTCGTTCCAGTGCATCCTCCATGGCATCAAGTAATTGGCGCAGC TCCATGAGTACCACGTCAAGTGCGGGGACGTCGACATCGGCATTCACGCACTACAGTAATGGTAGCACGCGCAGT ATTTCGACCACTGCAACCAGTGTCTCAGCTACCAGTTGGAGGACCAGTGTGAAGCAGCCATCGATCTCATCCAAC TCGAGCGGGTACTCAGCGTCCGGTGGCTATACTTCCGTCCCTAAGAACATCAAGCTCATGAATGGCGTGCCTTGG GAACTGGACCAGTTGCCGCGAGGACAACATCCCGATCCTTCGTTCGGTAACCCTCCCCCTCGCAAGCAGCGTACG AGGAAACACAAGGACATCAAACTTGACACCATCACTGAACGGCCATCCAACTCGGCGGGCCTGAAATCTCCTGAG CTCCGAAAGGACGCGATGACGAGCACGTCTGACCTCCCGGCGGTTGTAAATGCTGCCAGGGATGAGGGGGACAAT GTCGGTAGTGGTGGTGTGAAGAAGGTGCACAAGGGACAGATCAACGCGCTCGCGAAGATGTTGTCAGCCTTGCGA CGATGA |
| Length | 3156 |
Transcript
| Sequence id | CopciAB_471344.T0 |
|---|---|
| Sequence |
>CopciAB_471344.T0 ATCTTCTCCAGTCTCCCAACTTTTCGCCGCCGCCGTCGTTGTCGTCGTTGAACGTCGCTCTTCTCGTCGGCCTCT TCATCTACGCTTTTCTCGTTCCACAGCAAAGCTAGGCGTCGACGGGGCGTCCTCTCCCTTCTCCGACACGTTCTC AACCACGGTCTCCTGCATGCCCTCAACGTCCTCATAGTCTCTGACCCCGGTCGATTTCAACCGACACTCTACGTC CCCTGCGCTCAGCATCGCTGACAGCCACGAACCGTTTTTCAGCTGCCTCTGTCGCTCATGTTGCAGTACCCGGTT CAGGTTCCCTCGCTGTCCCTCTCCTCATCTGCAAAGCTCGCCTCTAAACCCTCTAAAGATAACATTCCTTATTTG CTCAGTAATGACCCCCCTCGCCAGCCACAGCCATTGAGCTCCCTCCGCTTCCAGAAGGACCCGTATGCTGCCAGG CCAGTTGGTCCAAATCGTTTGCCAGCACCGATCGTGCCACTCTCGATTTCGACCAATCTGGTCCCGTCACCGTCA GCTTCGTCCCCGTCAACCGCCTCGGCCAAGCACCCACGTAGCCCTTACATCTCCTCCCGTCAGCTTTCTGCAAGC CCCGTCCCGTTCCGCCAGCAACCCAGCGGCCCAGCACCCCCTTCGGCTCTATCCCAATGCGCCTCTTCAGGCTCG TATCCCATTGAAGACGTGTTAACACCTGGGGATATTGTCGGCGAGGGCCTTGTTCTCCAGAATGAGGTTCTCCGG TTGGTTGGGTCCACGAACGAGTCAGATGCAGCTGCACCAGCCAAGGAGTTTGAAGTCGTGCGTGTCCTCGGCACT GGCAGCTACGCTGTTGTCTACCTGGTTCGAGAAATTCTTTCCCGACCCCCTCCTTCTGAAGACGGCCACATGACA ACCATCGGCTCCATGGATATGGACGGCACCCCTCAGCTAGAGTATGGGCGTGAATACGCTATCAAATTGCTGTCC AAGGCCAACCTTGACGAGGAGGCTCTGGCCGCCCAAATGACTGAGGTTACGATCCATCAATCTCTTCCCCTCCAC CCAAATATTGTAACGCTACACCGCACACTTGAGACCCCTTCTTTCTTACTTCTTCTCCTGGAATACGTTCCGGGC GAGGACCTTTTCTATTTTTTGGAACAAGCCCGTGACCATTATGAACCCCAGCCTTCTGATTTTGTCATGTACAAC TCTCGCACTCCCCCAACTCCCAGTTTACTCTCCAATCTCCACCCTTCGCAACTGCTCTCGCGCACTCGCCTTCGC CTCATCGCTTCGATGTTCTCCCAGATGTGTGACGCTGTTGCCGCCTGCCACTCCACCCAAGTCTTCCACCGCGAT ATCAAGCCTGAAAACTTTATCGTTACAGATGGTTGGGTGATGCTGGCGAACGGACGTCAAGAACGAAAGGTCATT GTCAAGTTGACGGACTTCGGTTTGTCAACTACCGACATCGACTCTTCGGACGTCGATTGTGGAAGCGCGCCCTAC ATGAGCTACGAATGCAGGAACAACATTGCTCCCACCTATCGACCTCGCGATGCTGATGTTTGGTCTCTTGGCATC GTGCTAATCAACATGTTGTATCACTTTAACCCATGGACCGACACGGCCGAAGGTGCTTGCTCGTCGTTCGACCTT TACCGTCGGGATCCCGTCAATTTCTTCACCAATCGTTTCCCCGGAATGACTCGAGGTGTTGCTGAGTTCTTGGCT TCAAGAGTCTTTTGCATCCTCGAGGGTGATGAGAGCGAGGGCCGACGCATCTCTGCGGCCGATTTCGGTGTTTGG GTCAAGGATAACTTGGTGACGATGTTGGCTGGCCATGGGATGCAGCGCACTTTGTCCACCAGTTCGACGCAAGGT CATCCTATTACCTCCATCCCCGTGTCTCATCGTCCCATGTCCCGTAATCCCTCCAACGCGACCGCCGCTCGTGGT ACTCCTGCCATGCCCACTAGATCGCTTTCCCGGGCACCATCCCTCGGCCCCGCCTACGAACGCGAGGAGAAATCA GAGCTGTCCACTGTCATCGATCAAGATAACGAAGATGATGTCGAGGTGCCCGAGCCTTCTGAACCTATCCTCGGC GTCGAAGAAGCTGGCTCTCGATCTGCCTCCACTTCCAAGCGTCGTAAGCGCGGAGCTCGCAAAGGCAAAGGTGCC CAAGCTTCGTCGCCTACATCCCCCCAAGACGATACAATTGTCAACCTGGCCGCGGCTTCTGAGTGTTTGGCTCGG GAGCTGAGCAAGCAGTCGAAGGTCTCCCTTCCCTCTTCACCCAAGAGCACTTCCACCTCCTCGCGCAAGCGACAG CCCCTCGAACCCGTATCCATGTACGCCATGCCCACCCCTCTGCTTAATTCTTCTACCCCTCGAACTGCAACGAAA GCTTCAACATTTGTCACTTGCACTCCTGTACCACCCTCGGCTCCAACGAAGAAGCCATCCCGGTGGAAGCTCAGC TTTGGTAAGGCCAACGCATCAACCTTGGCAAACGCCGTCGAGCGGCCTGCGACTGAAGAAGCTGCATCACCAGCT TCTGCTGAACCATCTCAACCACCACCTCCGTCTACCACTGTCAACAACGTCACGAATCTTATCATGGGCTTGGAT GCTCCTGCTGCCCGTCCTATTCCTCCTGCTGCAGGTGTATTTGACGATGGTGCATCTACTTGGGCACGTGGCCGT CGACCAAGAGACGCCTCCCCTGTGCGCAAGTATGGTGGACGGTCTCCCCGACGAGGACTTCAATCTCCTTCCCAG CCCAGCTTGGCTGACAGTTGGGTTGACCCTCCTTCGCGCGGTCCTTCACGTGCCATCTCTCCTAGCAGTACGCGA AGCGGGCGTCCGATTCGTTCCAGTGCATCCTCCATGGCATCAAGTAATTGGCGCAGCTCCATGAGTACCACGTCA AGTGCGGGGACGTCGACATCGGCATTCACGCACTACAGTAATGGTAGCACGCGCAGTATTTCGACCACTGCAACC AGTGTCTCAGCTACCAGTTGGAGGACCAGTGTGAAGCAGCCATCGATCTCATCCAACTCGAGCGGGTACTCAGCG TCCGGTGGCTATACTTCCGTCCCTAAGAACATCAAGCTCATGAATGGCGTGCCTTGGGAACTGGACCAGTTGCCG CGAGGACAACATCCCGATCCTTCGTTCGGTAACCCTCCCCCTCGCAAGCAGCGTACGAGGAAACACAAGGACATC AAACTTGACACCATCACTGAACGGCCATCCAACTCGGCGGGCCTGAAATCTCCTGAGCTCCGAAAGGACGCGATG ACGAGCACGTCTGACCTCCCGGCGGTTGTAAATGCTGCCAGGGATGAGGGGGACAATGTCGGTAGTGGTGGTGTG AAGAAGGTGCACAAGGGACAGATCAACGCGCTCGCGAAGATGTTGTCAGCCTTGCGACGATGAAGAACGTCCCGT GTCGTTTTGTCTTTTGTGCCTTTCGCCCTGACGGACTGTTGTATCGAGACAGGAACTCTTCCGCTCCCTATGACT TCTTGTTGAACTGATGTTGCCAATGCGAGTAGTTTTGCGGACAATGCGTTTGGCCCCCCAATCCTTTTTGTGCGT GGTGCGGATCAGGAATTCTCGTGTTGCGCTGTATTCATAAAGTATCCTACTTCACGGTGCGCAGTCAATACTTGG ACATCCCCTCGTGTTATGGGTTGATACCCTGCATTCGTTTGTGACGGCGTTGATTCATCGTTTTAATCTCTCTGT TCTATCCAGTATTGTACTATCTTTCGACGGGCTCCGTGCAGCACCGCTGTCATTGTCATAACTCACACATTACAT TGATATCACATACATACGAAACATTCACTCCTTGCATTTTTTCTCTTTACTTGCTCTATACACTCAAGTCTACGC CTTGTAGTAGTTGTATTTAATATTCGAAGTATGGTCAATGGCCGCGAGCCCC |
| Length | 3952 |
Gene
| Sequence id | CopciAB_471344.T0 |
|---|---|
| Sequence |
>CopciAB_471344.T0 ATCTTCTCCAGTCTCCCAACTTTTCGCCGCCGCCGTCGTTGTCGTCGTTGAACGTCGCTCTTCTCGTCGGCCTCT TCATCTACGGTGCGTTGCTGCTTGTCGTCGTCGTTGTCGACGTCGACGGCGCCCCGTTTGCCACCGTCCCCAGAG TTGCTGATTCATTACCCCAGCTTTTCTCGTTCCACAGCAAAGCTAGGCGTCGACGGGGCGTCCTCTCCCTTCTCC GACACGTTCTCAACCACGGTCTCCTGCATGCCCTCAACGTCCTCATAGTCTCTGACCCCGGTCGATTTCAACCGA CACTCTACGTCCCCTGCGCTCAGCATCGCTGACAGCCACGAACCGTTTTTCAGCTGCCTCTGTCGCTCATGTTGC AGTACCCGGTTCAGGTTCCCTCGCTGTCCCTCTCCTCATCTGCAAAGCTCGCCTCTAAACCCTCTAAAGATAACA TTCCTTATTTGCTCAGTAATGACCCCCCTCGCCAGCCACAGCCATTGAGCTCCCTCCGCTTCCAGAAGGACCCGT ATGCTGCCAGGCCAGTTGGTCCAAATCGTTTGCCAGCACCGATCGTGCCACTCTCGATTTCGACCAATCTGGTCC CGTCACCGTCAGCTTCGTCCCCGTCAACCGCCTCGGCCAAGCACCCACGTAGCCCTTACATCTCCTCCCGTCAGC TTTCTGCAAGCCCCGTCCCGTTCCGCCAGCAACCCAGCGGCCCAGCACCCCCTTCGGCTCTATCCCAATGCGCCT CTTCAGGCTCGTATCCCATTGAAGACGTGTTAACACCTGGGGATATTGTCGGCGAGGGCCTTGTTCTCCAGAATG AGGTTCTCCGGTTGGTTGGGTCCACGAACGAGTCAGATGCAGCTGCACCAGCCAAGGAGTTTGAAGTCGTGCGTG TCCTCGGCACTGGCAGCTACGCTGTTGTCTACCTGGTTCGAGAAATTCTTTCCCGACCCCCTCCTTCTGAAGACG GCCACATGACAACCATCGGCTCCATGGATATGGACGGCACCCCTCAGCTAGAGTATGGGCGTGAATACGCTATCA AATTGCTGTCCAAGGCCAACCTTGACGAGGAGGCTCTGGCCGCCCAAATGACTGAGGTGCGTGTTTAGATCAGAA GCTCCGTGTGTTCAGTAGTTGACACCATGTGCAGGTTACGATCCATCAATCTCTTCCCCTCCACCCAAATATTGT AACGCTACACCGCACACTTGAGACCCCTTCTTTCTTACTTCTTCTCCTGGAATACGTTCCGGGCGAGGACCTTTT CTATTTTTTGGAACAAGCCCGTGACCATTATGAACCCCAGCCTTCTGATTTTGTCATGTACAACTCTCGCACTCC CCCAACTCCCAGTTTACTCTCCAATCTCCACCCTTCGCAACTGCTCTCGCGCACTCGCCTTCGCCTCATCGCTTC GATGTTCTCCCAGATGTGTGACGCTGTTGCCGCCTGCCACTCCACCCAAGTCTTCCACCGCGATATCAAGCCTGA AAACTTTATCGTTACAGATGGTTGGGTGATGCTGGCGAACGGACGTCAAGAACGAAAGGTCATTGTCAAGTTGAC GGACTTCGGTTTGTCAACTACCGACATCGACTCTTCGGACGTCGATTGTGGAAGCGCGCCCTACATGAGCTACGG TGAGCTCAACTTGACGTATCCATATTGTAGCAGTTGATACTAACACACATGGTCAGAATGCAGGAACAACATTGC TCCCACCTATCGACCTCGCGATGCTGATGTTTGGTCTCTTGGCATCGTGCTAATCAACATGTGCGGACTCGCTAC GACTTTCTTCCCTAATTCAGCGTGCTAACATATAATTCAGGTTGTATCACTTTAACCCATGGACCGACACGGCCG AAGGTGCTTGCTCGTCGTTCGACCTTTACCGTCGGGATCCCGTCAATTTCTTCACCAATCGTTTCCCCGGAATGA CTCGAGGTGTTGCTGAGTTCTTGGCTTCAAGAGTCTTTTGCATCCTCGAGGGTGATGAGAGCGAGGGCCGACGCA TCTCTGCGGCCGATTTCGGTGTTTGGGTCAAGGATAACTTGGTGACGATGTTGGCTGGCCATGGGATGCAGCGCA CTTTGTCCACCAGTTCGACGCAAGGTCATCCTATTACCTCCATCCCCGTGTCTCATCGTCCCATGTCCCGTAATC CCTCCAACGCGACCGCCGCTCGTGGTACTCCTGCCATGCCCACTAGATCGCTTTCCCGGGCACCATCCCTCGGCC CCGCCTACGAACGCGAGGAGAAATCAGAGCTGTCCACTGTCATCGATCAAGATAACGAAGATGATGTCGAGGTGC CCGAGCCTTCTGAACCTATCCTCGGCGTCGAAGAAGCTGGCTCTCGATCTGCCTCCACTTCCAAGCGTCGTAAGC GCGGAGCTCGCAAAGGCAAAGGTGCCCAAGCTTCGTCGCCTACATCCCCCCAAGACGATACAATTGTCAACCTGG CCGCGGCTTCTGAGTGTTTGGCTCGGGAGCTGAGCAAGCAGTCGAAGGTCTCCCTTCCCTCTTCACCCAAGAGCA CTTCCACCTCCTCGCGCAAGCGACAGCCCCTCGAACCCGTATCCATGTACGCCATGCCCACCCCTCTGCTTAATT CTTCTACCCCTCGAACTGCAACGAAAGCTTCAACATTTGTCACTTGCACTCCTGTACCACCCTCGGCTCCAACGA AGAAGCCATCCCGGTGGAAGCTCAGCTTTGGTAAGGCCAACGCATCAACCTTGGCAAACGCCGTCGAGCGGCCTG CGACTGAAGAAGCTGCATCACCAGCTTCTGCTGAACCATCTCAACCACCACCTCCGTCTACCACTGTCAACAACG TCACGAATCTTATCATGGGCTTGGATGCTCCTGCTGCCCGTCCTATTCCTCCTGCTGCAGGTGTATTTGACGATG GTGCATCTACTTGGGCACGTGGCCGTCGACCAAGAGACGCCTCCCCTGTGCGCAAGTATGGTGGACGGTCTCCCC GACGAGGACTTCAATCTCCTTCCCAGCCCAGCTTGGCTGACAGTTGGGTTGACCCTCCTTCGCGCGGTCCTTCAC GTGCCATCTCTCCTAGCAGTACGCGAAGCGGGCGTCCGATTCGTTCCAGTGCATCCTCCATGGCATCAAGTAATT GGCGCAGCTCCATGAGTACCACGTCAAGTGCGGGGACGTCGACATCGGCATTCACGCACTACAGTAATGGTAGCA CGCGCAGTATTTCGACCACTGCAACCAGTGTCTCAGCTACCAGTTGGAGGACCAGTGTGAAGCAGCCATCGATCT CATCCAACTCGAGCGGGTACTCAGCGTCCGGTGGCTATACTTCCGTCCCTAAGAACATCAAGCGTAAGTCCTTGA ATTGACTCGTGAAGGAAATTACCCTGATTCATGTCTTTTCAGTCATGAATGGCGTGCCTTGGGAACTGGACCAGT TGCCGCGAGGACAACATCCCGATCCTTCGTTCGGTAACCCTCCCCCTCGCAAGCAGCGTACGAGGAAACACAAGG ACATCAAACTTGACACCATCACTGAACGGCCATCCAACTCGGCGGGCCTGAAATCTCCTGAGCTCCGAAAGGACG CGATGACGAGCACGTCTGACCTCCCGGCGGTTGTAAATGCTGCCAGGGATGAGGGGGACAATGTCGGTAGTGGTG GTGTGAAGAAGGTGCACAAGGGACAGATCAACGCGCTCGCGAAGATGTTGTCAGCCTTGCGACGATGAAGAACGT CCCGTGTCGTTTTGTCTTTTGTGCCTTTCGCCCTGACGGACTGTTGTATCGAGACAGGAACTCTTCCGCTCCCTA TGACTTCTTGTTGAACTGATGTTGCCAATGCGAGTAGTTTTGCGGACAATGCGTTTGGCCCCCCAATCCTTTTTG TGCGTGGTGCGGATCAGGAATTCTCGTGTTGCGCTGTATTCATAAAGTATCCTACTTCACGGTGCGCAGTCAATA CTTGGACATCCCCTCGTGTTATGGGTTGATACCCTGCATTCGTTTGTGACGGCGTTGATTCATCGTTTTAATCTC TCTGTTCTATCCAGTATTGTACTATCTTTCGACGGGCTCCGTGCAGCACCGCTGTCATTGTCATAACTCACACAT TACATTGATATCACATACATACGAAACATTCACTCCTTGCATTTTTTCTCTTTACTTGCTCTATACACTCAAGTC TACGCCTTGTAGTAGTTGTATTTAATATTCGAAGTATGGTCAATGGCCGCGAGCCCC |
| Length | 4257 |