CopciAB_473299
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_473299 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 473299 |
| Uniprot id | Functional description | ||
| Location | scaffold_1:2310501..2312818 | Strand | - |
| Gene length (nt) | 2318 | Transcript length (nt) | 2143 |
| CDS length (nt) | 2019 | Protein length (aa) | 672 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Lentinula edodes B17 | Lened_B_1_1_2113 | 31.4 | 1.984E-53 | 201 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_13509 | 31.4 | 2.489E-53 | 201 |
| Lentinula edodes NBRC 111202 | Lenedo1_258749 | 32.5 | 2.04E-50 | 192 |
| Schizophyllum commune H4-8 | Schco3_2698233 | 30.5 | 1.241E-43 | 171 |
| Grifola frondosa | Grifr_OBZ79138 | 29.9 | 2.402E-38 | 154 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_113461 | 28.4 | 8.949E-37 | 149 |
| Pleurotus eryngii ATCC 90797 | Pleery1_69230 | 29.1 | 7.422E-35 | 143 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29262 | 24.8 | 2.7E-32 | 135 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_473299.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| No records | |||||
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
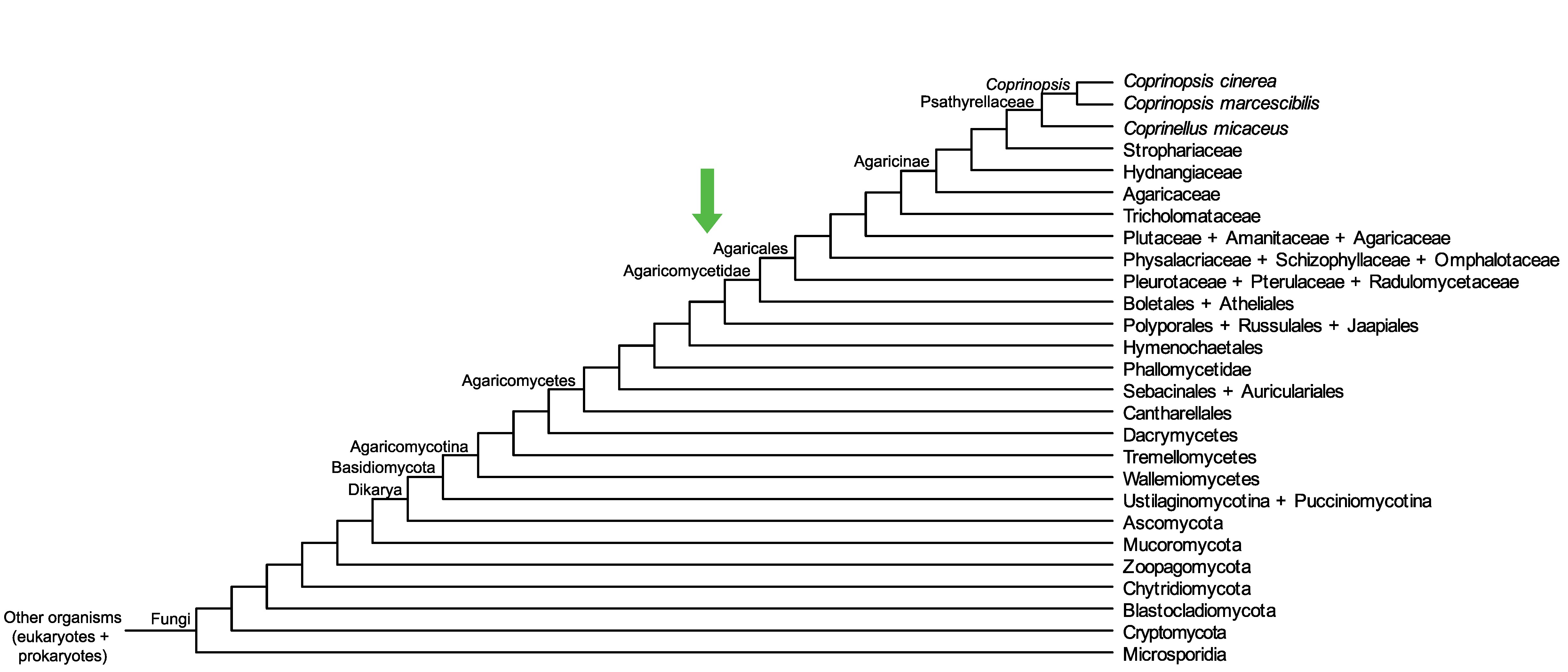
Conservation of CopciAB_473299 across fungi.
Arrow shows the origin of gene family containing CopciAB_473299.
Protein
| Sequence id | CopciAB_473299.T0 |
|---|---|
| Sequence |
>CopciAB_473299.T0 MRETRTGPLAKIRSFINDSGILTMAPFKLRRLGTMAHHNKSESFEPLETVSEWLPKSKFFKALTTPFDYIPMESL KSLKRNSNGEACPPMASHIKTARGRGHYSIPIWTSGGPWKWAQGLPLVDSRIIFAAEPHPFNHTKEWYGLGFLGD ELLQFMCRTWDFQSEGSMARFIADVALNKALLYPLHGKYVSTIIGIYAGNETAHVVTEVPHRRFWVAASPDMPEA LKRRCIEAYEHLHSRGILHGNVGLDKIAIGGDGRVTLLDFSQAKAIEPVRDCYMAMANPGDLRMELRKVKCLLNF QDAVEKERDKHNWQYHRTCWNSEEQEKQDIDPNYVPDLQDIPEEERLDPPVDVDLEGWVAGPDYIPDRFVVPGTS PEQYAIAVGEFYRLALEMEEKDKTPTPSPSSSSFSSGSTAPSPSWTLRQAAAQNLQATEQPSLAETTAESRKRKA GSTESESETPSKRMRTLGAHETVAAGPSRIVAESSHSPGILKVTASIHPSIMRNNLDHGMPMIKVIDFAHHIPPP RARVPNSLEEKDSPSTRSPRSSSENRRPPQKRVAFSAGRDTAFKRYKVPRSMWHYSDIQLRRETNKRLRRIGSGP RQKSLALQAYELRKKVLISHPVERRISARLQAKRERERRPSVVESPSTGPTTASLQAFWRWLSDTGSRLTTY |
| Length | 672 |
Coding
| Sequence id | CopciAB_473299.T0 |
|---|---|
| Sequence |
>CopciAB_473299.T0 ATGCGGGAAACAAGGACTGGCCCTTTGGCCAAAATTCGATCATTTATAAACGACAGTGGTATCTTGACTATGGCC CCCTTCAAACTACGACGACTTGGAACCATGGCCCATCACAACAAATCGGAATCGTTCGAACCTCTAGAGACGGTG TCAGAATGGCTACCTAAATCCAAATTCTTCAAAGCGCTGACCACACCATTCGACTACATACCCATGGAATCCTTG AAATCTCTGAAACGAAATTCCAATGGCGAGGCCTGCCCCCCAATGGCCTCTCATATCAAGACAGCTCGCGGCAGG GGGCACTATTCCATCCCTATCTGGACTAGCGGAGGACCTTGGAAATGGGCACAGGGCCTCCCTCTTGTCGACAGT CGAATCATTTTTGCAGCCGAACCCCACCCTTTCAATCACACCAAGGAGTGGTATGGCCTGGGATTTCTCGGAGAT GAGCTCCTTCAATTTATGTGTAGGACGTGGGATTTCCAGTCAGAAGGCTCTATGGCTAGGTTCATTGCCGACGTG GCGTTGAACAAAGCCCTTTTGTACCCTCTTCACGGAAAATACGTCTCGACTATCATCGGCATCTACGCTGGAAAC GAAACTGCGCATGTTGTTACAGAAGTCCCACATCGACGATTCTGGGTTGCAGCATCTCCCGACATGCCAGAAGCC CTGAAGAGACGCTGTATCGAAGCCTATGAGCACCTACACTCCCGTGGGATTCTTCATGGTAACGTTGGTTTGGAC AAAATCGCCATTGGGGGAGATGGCCGGGTCACGCTCCTCGACTTCAGTCAAGCCAAAGCCATCGAGCCCGTTCGG GACTGTTATATGGCGATGGCAAACCCAGGGGATTTGCGGATGGAGCTAAGAAAGGTGAAATGCCTGCTGAACTTC CAGGATGCCGTGGAGAAGGAGAGGGACAAACACAATTGGCAATACCATAGGACCTGCTGGAATTCCGAAGAGCAA GAAAAACAGGACATCGATCCAAACTATGTTCCCGACCTCCAAGATATCCCAGAAGAGGAACGCCTCGATCCACCT GTCGACGTCGACCTCGAGGGGTGGGTGGCTGGCCCAGACTACATCCCGGACAGGTTTGTTGTACCTGGAACATCT CCTGAACAGTATGCCATTGCCGTTGGAGAGTTTTACCGCCTGGCCCTGGAAATGGAGGAGAAGGACAAAACACCA ACCCCTTCCCCTTCCTCTTCCTCATTTTCGTCTGGGAGCACGGCCCCTAGTCCCTCCTGGACATTGCGTCAGGCG GCAGCACAGAATCTGCAGGCAACGGAGCAACCCAGTCTAGCTGAAACGACTGCCGAGTCCCGAAAGCGCAAGGCC GGCTCCACAGAAAGCGAATCCGAAACCCCATCAAAAAGAATGCGGACTCTGGGAGCCCATGAAACCGTTGCAGCG GGACCAAGTCGGATAGTGGCTGAATCAAGTCACTCTCCCGGTATCCTCAAAGTTACGGCCAGTATCCATCCTTCG ATAATGCGTAACAATCTCGACCACGGAATGCCCATGATCAAGGTCATAGACTTTGCACATCACATCCCTCCCCCT CGCGCCCGAGTCCCTAATTCCCTGGAGGAAAAAGACTCGCCTTCAACACGATCCCCCCGTTCATCATCAGAGAAT CGCAGGCCCCCTCAGAAAAGAGTTGCGTTCTCTGCGGGACGTGATACCGCGTTCAAGCGCTACAAGGTTCCACGA TCGATGTGGCATTACTCAGACATCCAGCTGCGACGAGAGACCAACAAGAGGCTTCGGCGAATTGGGAGTGGCCCG AGGCAAAAATCACTCGCACTCCAAGCTTACGAGCTGCGGAAGAAAGTTCTGATATCCCACCCTGTCGAGCGCCGC ATATCTGCAAGGCTACAGGCAAAGCGGGAGCGGGAAAGACGGCCATCCGTAGTAGAATCTCCTTCGACAGGCCCG ACTACAGCATCCTTGCAAGCATTCTGGAGGTGGTTGAGTGACACGGGCTCCAGGCTTACGACGTATTGA |
| Length | 2019 |
Transcript
| Sequence id | CopciAB_473299.T0 |
|---|---|
| Sequence |
>CopciAB_473299.T0 AGCTTGTTGTTCTCATTCTCGGCTCTGGTCAATGCGGGAAACAAGGACTGGCCCTTTGGCCAAAATTCGATCATT TATAAACGACAGTGGTATCTTGACTATGGCCCCCTTCAAACTACGACGACTTGGAACCATGGCCCATCACAACAA ATCGGAATCGTTCGAACCTCTAGAGACGGTGTCAGAATGGCTACCTAAATCCAAATTCTTCAAAGCGCTGACCAC ACCATTCGACTACATACCCATGGAATCCTTGAAATCTCTGAAACGAAATTCCAATGGCGAGGCCTGCCCCCCAAT GGCCTCTCATATCAAGACAGCTCGCGGCAGGGGGCACTATTCCATCCCTATCTGGACTAGCGGAGGACCTTGGAA ATGGGCACAGGGCCTCCCTCTTGTCGACAGTCGAATCATTTTTGCAGCCGAACCCCACCCTTTCAATCACACCAA GGAGTGGTATGGCCTGGGATTTCTCGGAGATGAGCTCCTTCAATTTATGTGTAGGACGTGGGATTTCCAGTCAGA AGGCTCTATGGCTAGGTTCATTGCCGACGTGGCGTTGAACAAAGCCCTTTTGTACCCTCTTCACGGAAAATACGT CTCGACTATCATCGGCATCTACGCTGGAAACGAAACTGCGCATGTTGTTACAGAAGTCCCACATCGACGATTCTG GGTTGCAGCATCTCCCGACATGCCAGAAGCCCTGAAGAGACGCTGTATCGAAGCCTATGAGCACCTACACTCCCG TGGGATTCTTCATGGTAACGTTGGTTTGGACAAAATCGCCATTGGGGGAGATGGCCGGGTCACGCTCCTCGACTT CAGTCAAGCCAAAGCCATCGAGCCCGTTCGGGACTGTTATATGGCGATGGCAAACCCAGGGGATTTGCGGATGGA GCTAAGAAAGGTGAAATGCCTGCTGAACTTCCAGGATGCCGTGGAGAAGGAGAGGGACAAACACAATTGGCAATA CCATAGGACCTGCTGGAATTCCGAAGAGCAAGAAAAACAGGACATCGATCCAAACTATGTTCCCGACCTCCAAGA TATCCCAGAAGAGGAACGCCTCGATCCACCTGTCGACGTCGACCTCGAGGGGTGGGTGGCTGGCCCAGACTACAT CCCGGACAGGTTTGTTGTACCTGGAACATCTCCTGAACAGTATGCCATTGCCGTTGGAGAGTTTTACCGCCTGGC CCTGGAAATGGAGGAGAAGGACAAAACACCAACCCCTTCCCCTTCCTCTTCCTCATTTTCGTCTGGGAGCACGGC CCCTAGTCCCTCCTGGACATTGCGTCAGGCGGCAGCACAGAATCTGCAGGCAACGGAGCAACCCAGTCTAGCTGA AACGACTGCCGAGTCCCGAAAGCGCAAGGCCGGCTCCACAGAAAGCGAATCCGAAACCCCATCAAAAAGAATGCG GACTCTGGGAGCCCATGAAACCGTTGCAGCGGGACCAAGTCGGATAGTGGCTGAATCAAGTCACTCTCCCGGTAT CCTCAAAGTTACGGCCAGTATCCATCCTTCGATAATGCGTAACAATCTCGACCACGGAATGCCCATGATCAAGGT CATAGACTTTGCACATCACATCCCTCCCCCTCGCGCCCGAGTCCCTAATTCCCTGGAGGAAAAAGACTCGCCTTC AACACGATCCCCCCGTTCATCATCAGAGAATCGCAGGCCCCCTCAGAAAAGAGTTGCGTTCTCTGCGGGACGTGA TACCGCGTTCAAGCGCTACAAGGTTCCACGATCGATGTGGCATTACTCAGACATCCAGCTGCGACGAGAGACCAA CAAGAGGCTTCGGCGAATTGGGAGTGGCCCGAGGCAAAAATCACTCGCACTCCAAGCTTACGAGCTGCGGAAGAA AGTTCTGATATCCCACCCTGTCGAGCGCCGCATATCTGCAAGGCTACAGGCAAAGCGGGAGCGGGAAAGACGGCC ATCCGTAGTAGAATCTCCTTCGACAGGCCCGACTACAGCATCCTTGCAAGCATTCTGGAGGTGGTTGAGTGACAC GGGCTCCAGGCTTACGACGTATTGACACGAATTCGGCCCGCTACACCTACTCCGTTTGCTCTGTCACTTCAGCAA CTTCCACGTCAAATATGCGCTAAATATATAGTCCTGGATGTCA |
| Length | 2143 |
Gene
| Sequence id | CopciAB_473299.T0 |
|---|---|
| Sequence |
>CopciAB_473299.T0 AGCTTGTTGTTCTCATTCTCGGCTCTGGTCAATGCGGGAAACAAGGACTGGCCCTTTGGCCAAAATTCGATCATT TATAAACGACAGTGGTATCTTGACTATGGCCCCCTTCAAACTACGACGACTTGGAACCATGGCCCATCACAACAA ATCGGAATCGTTCGAACCTCTAGAGACGGTGTCAGAATGGCTACCTAAATCCAAATTCTTCAAAGCGCTGACCAC ACCATTCGACTACATACCCATGGAATCCTTGAAATCTCTGAAACGAAATTCCAATGGCGAGGCCTGCCCCCCAAT GGCCTCTCATATCAAGACAGCTCGCGGCAGGGGGCACTATTCCATCCCTATCTGGACTAGCGGAGGACCTTGGAA ATGGGCACAGGGCCTCCCTCTTGTCGACAGTCGAATCATTTTTGCAGCCGAACCCCGTACGCCTATTTATTATCT CGCGCCTGTCCAACTAACTCCCTAGTTTCGATGTAGACCCTTTCAATCACACCAAGGAGTGGTATGGCCTGGGAT TTCTCGGAGATGAGCTCCTTCAATTTATGTGTAGGACGTACGTTTCGTTTCGTTCTCTCATAACAAGCAAAATTC CCTCACGCTGAGAGCAGGTGGGATTTCCAGTCAGAAGGCTCTATGGCTAGGTTCATTGCCGACGTGGCGTTGAAC AAAGCCCTTTTGTACCCTCTTCACGGAAAATACGTCTCGACTATCATCGGCATCTACGCTGGAAACGAAACTGCG CATGTTGTTACAGAAGTCCCACATCGACGATTCTGGGTTGCAGCATCTCCCGACATGCCAGAAGCCCTGAAGAGA CGCTGTATCGAAGCCTATGAGCACCTACACTCCCGTGGGATTCTTCATGGTAACGTTGGTTTGGACAAAATCGCC ATTGGGGGAGATGGCCGGGTCACGCTCCTCGACTTCAGTCAAGCCAAAGCCATCGAGCCCGTTCGGGACTGTTAT ATGGCGATGGCAAACCCAGGGGATTTGCGGATGGAGCTAAGAAAGGTGAAATGCCTGCTGAACTTCCAGGATGCC GTGGAGAAGGAGAGGGACAAACACAATTGGCAATACCATAGGACCTGCTGGAATTCCGAAGAGCAAGAAAAACAG GACATCGATCCAAACTATGTTCCCGACCTCCAAGATATCCCAGAAGAGGAACGCCTCGATCCACCTGTCGACGTC GACCTCGAGGGGTGGGTGGCTGGCCCAGACTACATCCCGGACAGGTTTGTTGTACCTGGAACATCTCCTGAACAG TATGCCATTGCCGTTGGAGAGTTTTACCGCCTGGCCCTGGAAATGGAGGAGAAGGACAAAACACCAACCCCTTCC CCTTCCTCTTCCTCATTTTCGTCTGGGAGCACGGCCCCTAGTCCCTCCTGGACATTGCGTCAGGCGGCAGCACAG AATCTGCAGGCAACGGAGCAACCCAGTCTAGCTGAAACGACTGCCGAGTCCCGAAAGCGCAAGGCCGGCTCCACA GAAAGCGAATCCGAAACCCCATCAAAAAGAATGCGGACTCTGGGAGCCCATGAAACCGTTGCAGCGGGACCAAGT CGGATAGTGGCTGAATCAAGTCACTCTCCCGGTATCCTCAAAGTTACGGCCAGTATCCGTACGCTGTCTCCTTTT CTTCTTTGGCGCCTTTTCGCTCAGTCCTTTACTACATTTGTGACGTAGATCCTTCGATAATGCGTAACAATCTCG ACCACGGAATGCCCATGATCAAGGTCATAGACTTTGCACATCACATCCCTCCCCCTCGCGCCCGAGTCCCTAATT CCCTGGAGGAAAAAGACTCGCCTTCAACACGATCCCCCCGTTCATCATCAGAGAATCGCAGGCCCCCTCAGAAAA GAGTTGCGTTCTCTGCGGGACGTGATACCGCGTTCAAGCGCTACAAGGTTCCACGATCGATGTGGCATTACTCAG ACATCCAGCTGCGACGAGAGACCAACAAGAGGCTTCGGCGAATTGGGAGTGGCCCGAGGCAAAAATCACTCGCAC TCCAAGCTTACGAGCTGCGGAAGAAAGTTCTGATATCCCACCCTGTCGAGCGCCGCATATCTGCAAGGCTACAGG CAAAGCGGGAGCGGGAAAGACGGCCATCCGTAGTAGAATCTCCTTCGACAGGCCCGACTACAGCATCCTTGCAAG CATTCTGGAGGTGGTTGAGTGACACGGGCTCCAGGCTTACGACGTATTGACACGAATTCGGCCCGCTACACCTAC TCCGTTTGCTCTGTCACTTCAGCAACTTCCACGTCAAATATGCGCTAAATATATAGTCCTGGATGTCA |
| Length | 2318 |