CopciAB_481308
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_481308 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | exgD | Synonyms | 481308 |
| Uniprot id | Functional description | Exo-beta-1,3-glucanase | |
| Location | scaffold_4:667285..670313 | Strand | + |
| Gene length (nt) | 3029 | Transcript length (nt) | 2522 |
| CDS length (nt) | 2163 | Protein length (aa) | 720 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB20695 | 56.8 | 1.339E-276 | 858 |
| Agrocybe aegerita | Agrae_CAA7260371 | 58.2 | 2.635E-276 | 857 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205212 | 57.2 | 1.153E-253 | 791 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_72198 | 57 | 1.201E-253 | 791 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_3948 | 53.6 | 3.714E-252 | 787 |
| Lentinula edodes B17 | Lened_B_1_1_1999 | 53.8 | 6.557E-252 | 786 |
| Lentinula edodes NBRC 111202 | Lenedo1_1057812 | 53.8 | 8.287E-252 | 786 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1050544 | 53.5 | 2.177E-247 | 773 |
| Pleurotus ostreatus PC9 | PleosPC9_1_117109 | 54.1 | 2.717E-244 | 764 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1403196 | 52.9 | 7.832E-243 | 760 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_121355 | 52 | 6.126E-235 | 737 |
| Flammulina velutipes | Flave_chr05AA00514 | 52.7 | 2.537E-216 | 683 |
| Schizophyllum commune H4-8 | Schco3_2645630 | 48.9 | 2.415E-209 | 663 |
| Grifola frondosa | Grifr_OBZ65976 | 47.3 | 1.221E-160 | 521 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | exgD |
|---|---|
| Protein id | CopciAB_481308.T0 |
| Description | Exo-beta-1,3-glucanase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 265 | 420 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 96 | 118 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Exo-beta-1,3-glucanase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_9 |
Transcription factor
| Group |
|---|
| No records |
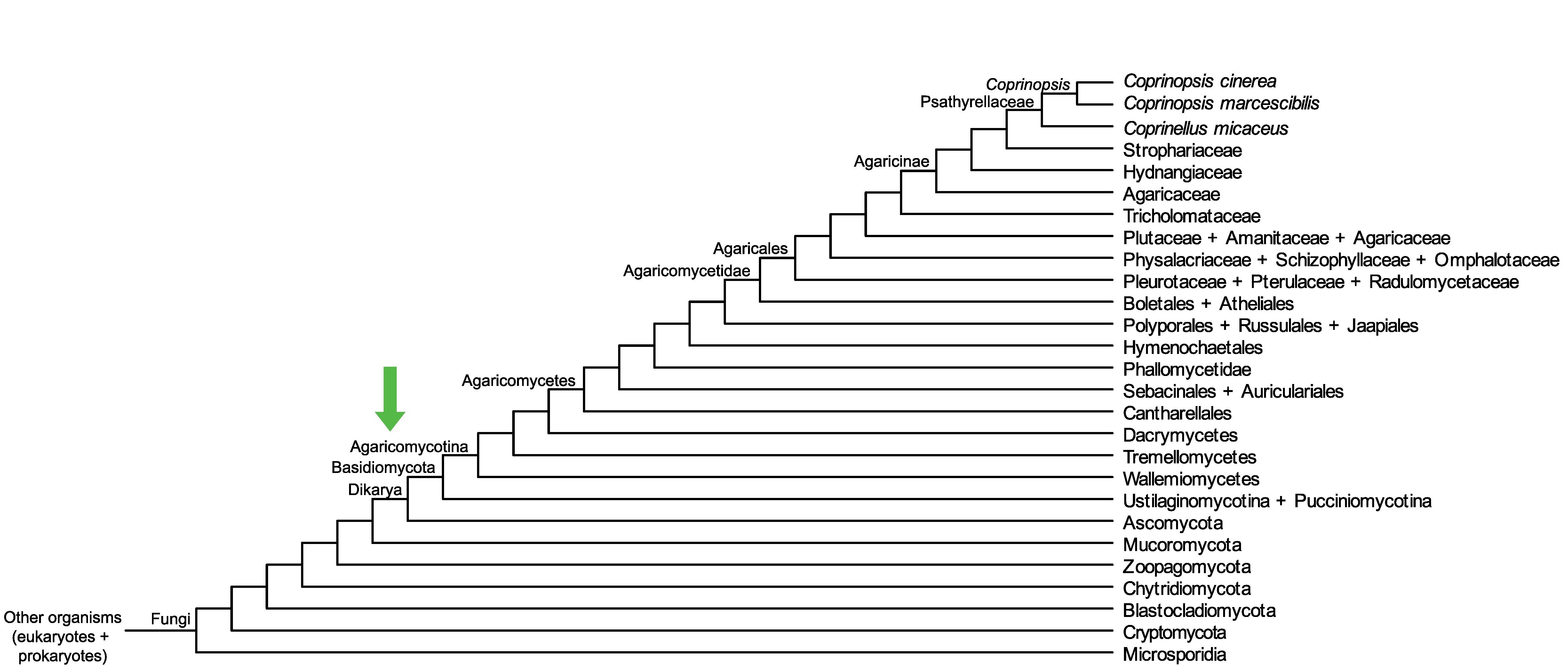
Conservation of CopciAB_481308 across fungi.
Arrow shows the origin of gene family containing CopciAB_481308.
Protein
| Sequence id | CopciAB_481308.T0 |
|---|---|
| Sequence |
>CopciAB_481308.T0 MAERPVSIVSDGALANAASVPLPQGDGNSLNDPQAALDSRTPSSAALRDSAALADSTTALGPGNNAVDSSKEAAD SAAEGGQAGEPASTSRKRRPLLIALLALLALAIVVLAVVLPVYFKVIKPRQNRDGSTDGSSGSGTDGNESTSAPS PTGSATPTNPTSGGDGSTVTTEDGTTFTYSNQFGGFWYSDPDDPYNDSAQPNSWTPPLNASFDYTTTRIFGVNLG GLFVLEPFISPALFQRYPGARDEWDLSVAMAADTANGGLDQLEEHYRTFITEQDIAEIAGAGLNWVRLPVPFWAI DKWPGEPFLERTSWRYIVRVLQWCRKYGLRVNLDLHTIPGSHNAYNHGGKLNAFNFLNGAMGMANAQRALYYIQV FTEFINQPEWRNVVPMFSIMNEPIIGTIGVDQLRSFYVEAHRIMREITGYGEGNGPYMVIHDSFRGPGPWAGFMT GADRVGMDVHPYFAFNGDSDPPTIDGGVGPGAGNGWPLRACNRFNAMMNDSRRDFGVTVAGEFSNAWQDCSLFLR GVGGRADYGGDCTPWLDSSGWSDGVKAGLLAFASAQMDGLGDWFFWTWKIGESERGIVESPFWSYQLGLRNGWMP TDPRTVIGTCQALGVYTPWDGTFQPYMIGGPGAGAPSDASSYPWPPTALGDGNAVAALPTYATTGSPVVLPAPTF TDSQGNEILPSGTQASHPAPTPIAGCTYPDGWDSDGVPPPALCGV |
| Length | 720 |
Coding
| Sequence id | CopciAB_481308.T0 |
|---|---|
| Sequence |
>CopciAB_481308.T0 ATGGCAGAGCGACCAGTTTCGATAGTGTCGGACGGCGCATTGGCGAATGCAGCATCGGTCCCTCTCCCTCAAGGA GATGGGAACTCGCTCAACGACCCTCAAGCAGCGCTCGACTCGCGGACGCCCTCCTCTGCTGCCTTGCGCGACTCC GCTGCACTAGCGGATTCAACTACAGCGCTCGGCCCCGGCAATAATGCAGTGGATTCATCGAAAGAAGCGGCGGAT TCAGCAGCGGAAGGTGGCCAGGCTGGTGAGCCTGCCTCCACCTCGAGGAAACGACGACCCCTTCTCATTGCCCTC CTGGCCCTGCTCGCGCTCGCAATTGTGGTGCTGGCCGTTGTCCTCCCGGTCTATTTCAAGGTCATCAAACCCCGA CAAAATCGAGATGGCTCGACCGATGGGTCCAGCGGCAGCGGCACCGACGGTAACGAATCGACATCAGCCCCTTCG CCTACCGGGTCTGCTACTCCCACCAACCCTACGTCTGGTGGTGACGGATCGACAGTCACGACCGAAGATGGAACT ACGTTTACCTATTCCAACCAGTTTGGCGGTTTCTGGTATTCGGATCCCGACGATCCTTACAATGACAGCGCCCAA CCAAACTCGTGGACACCTCCGCTGAACGCTAGCTTCGATTACACGACGACTCGCATCTTTGGCGTGAATCTTGGT GGTCTCTTCGTATTGGAACCTTTTATCTCCCCTGCTCTATTCCAGAGATACCCTGGTGCCAGAGATGAATGGGAT CTCAGTGTAGCTATGGCCGCAGATACCGCGAACGGCGGTCTCGATCAACTGGAGGAACATTACAGGACATTCATC ACGGAACAAGACATCGCTGAGATTGCGGGCGCCGGCTTGAACTGGGTCCGTCTCCCTGTTCCTTTCTGGGCGATA GACAAGTGGCCTGGTGAGCCCTTCCTTGAGAGGACTTCCTGGAGATATATCGTTCGAGTGCTGCAGTGGTGTCGA AAATACGGTCTCCGTGTGAACCTCGATCTTCACACTATCCCTGGTTCACACAATGCTTACAACCACGGCGGAAAA CTGAATGCGTTCAATTTCCTCAACGGCGCGATGGGCATGGCAAACGCCCAGAGGGCTTTGTACTATATCCAGGTA TTCACCGAGTTCATCAACCAGCCTGAATGGCGAAACGTCGTACCGATGTTCAGCATCATGAATGAGCCTATCATT GGCACCATCGGTGTTGATCAATTGCGTTCGTTCTACGTCGAAGCGCATCGCATTATGCGCGAAATTACTGGATAC GGAGAAGGCAATGGACCATACATGGTCATCCACGATAGTTTCCGTGGTCCTGGTCCGTGGGCTGGCTTCATGACT GGCGCTGATCGAGTGGGAATGGACGTTCACCCTTACTTCGCCTTCAATGGTGATTCCGACCCTCCGACTATCGAC GGTGGTGTTGGTCCTGGTGCAGGCAACGGTTGGCCTCTTCGCGCATGTAATCGATTCAATGCCATGATGAATGAC AGTCGAAGGGATTTCGGTGTTACAGTCGCTGGGGAATTCAGCAACGCCTGGCAAGATTGCTCTCTCTTCCTTCGT GGAGTAGGTGGGAGGGCGGACTACGGTGGTGACTGCACCCCCTGGCTGGACTCTTCCGGTTGGTCGGATGGTGTC AAGGCCGGCCTTTTGGCGTTTGCTTCTGCTCAAATGGATGGGCTTGGCGATTGGTTCTTCTGGACATGGAAGATT GGCGAATCAGAGCGCGGTATTGTTGAATCGCCCTTCTGGTCCTACCAGCTTGGTCTCCGAAATGGATGGATGCCG ACCGACCCTCGCACTGTAATTGGTACTTGCCAGGCCCTTGGTGTCTATACCCCATGGGATGGAACCTTCCAACCA TACATGATTGGAGGTCCTGGTGCAGGGGCACCTTCCGATGCTTCCAGCTATCCATGGCCACCCACCGCGCTCGGT GATGGCAATGCTGTCGCAGCACTTCCCACTTATGCTACCACCGGCAGTCCTGTCGTCCTGCCCGCACCCACTTTC ACCGATTCGCAAGGCAACGAGATCCTTCCTAGCGGCACTCAGGCAAGCCACCCCGCGCCAACACCCATCGCCGGA TGCACCTATCCTGATGGCTGGGACTCTGACGGTGTACCACCCCCTGCGCTTTGTGGAGTGTAA |
| Length | 2163 |
Transcript
| Sequence id | CopciAB_481308.T0 |
|---|---|
| Sequence |
>CopciAB_481308.T0 AGAGTGCACTTGCGGTCTATAATTCACCATGGTTGGCAGGTGGCAAGCGGATTGAGCGCCTGCTTCCCTTTCTTC CCTCTCCCTCCCTCCCTCCTCGACTGGTCCCATGGCAGAGCGACCAGTTTCGATAGTGTCGGACGGCGCATTGGC GAATGCAGCATCGGTCCCTCTCCCTCAAGGAGATGGGAACTCGCTCAACGACCCTCAAGCAGCGCTCGACTCGCG GACGCCCTCCTCTGCTGCCTTGCGCGACTCCGCTGCACTAGCGGATTCAACTACAGCGCTCGGCCCCGGCAATAA TGCAGTGGATTCATCGAAAGAAGCGGCGGATTCAGCAGCGGAAGGTGGCCAGGCTGGTGAGCCTGCCTCCACCTC GAGGAAACGACGACCCCTTCTCATTGCCCTCCTGGCCCTGCTCGCGCTCGCAATTGTGGTGCTGGCCGTTGTCCT CCCGGTCTATTTCAAGGTCATCAAACCCCGACAAAATCGAGATGGCTCGACCGATGGGTCCAGCGGCAGCGGCAC CGACGGTAACGAATCGACATCAGCCCCTTCGCCTACCGGGTCTGCTACTCCCACCAACCCTACGTCTGGTGGTGA CGGATCGACAGTCACGACCGAAGATGGAACTACGTTTACCTATTCCAACCAGTTTGGCGGTTTCTGGTATTCGGA TCCCGACGATCCTTACAATGACAGCGCCCAACCAAACTCGTGGACACCTCCGCTGAACGCTAGCTTCGATTACAC GACGACTCGCATCTTTGGCGTGAATCTTGGTGGTCTCTTCGTATTGGAACCTTTTATCTCCCCTGCTCTATTCCA GAGATACCCTGGTGCCAGAGATGAATGGGATCTCAGTGTAGCTATGGCCGCAGATACCGCGAACGGCGGTCTCGA TCAACTGGAGGAACATTACAGGACATTCATCACGGAACAAGACATCGCTGAGATTGCGGGCGCCGGCTTGAACTG GGTCCGTCTCCCTGTTCCTTTCTGGGCGATAGACAAGTGGCCTGGTGAGCCCTTCCTTGAGAGGACTTCCTGGAG ATATATCGTTCGAGTGCTGCAGTGGTGTCGAAAATACGGTCTCCGTGTGAACCTCGATCTTCACACTATCCCTGG TTCACACAATGCTTACAACCACGGCGGAAAACTGAATGCGTTCAATTTCCTCAACGGCGCGATGGGCATGGCAAA CGCCCAGAGGGCTTTGTACTATATCCAGGTATTCACCGAGTTCATCAACCAGCCTGAATGGCGAAACGTCGTACC GATGTTCAGCATCATGAATGAGCCTATCATTGGCACCATCGGTGTTGATCAATTGCGTTCGTTCTACGTCGAAGC GCATCGCATTATGCGCGAAATTACTGGATACGGAGAAGGCAATGGACCATACATGGTCATCCACGATAGTTTCCG TGGTCCTGGTCCGTGGGCTGGCTTCATGACTGGCGCTGATCGAGTGGGAATGGACGTTCACCCTTACTTCGCCTT CAATGGTGATTCCGACCCTCCGACTATCGACGGTGGTGTTGGTCCTGGTGCAGGCAACGGTTGGCCTCTTCGCGC ATGTAATCGATTCAATGCCATGATGAATGACAGTCGAAGGGATTTCGGTGTTACAGTCGCTGGGGAATTCAGCAA CGCCTGGCAAGATTGCTCTCTCTTCCTTCGTGGAGTAGGTGGGAGGGCGGACTACGGTGGTGACTGCACCCCCTG GCTGGACTCTTCCGGTTGGTCGGATGGTGTCAAGGCCGGCCTTTTGGCGTTTGCTTCTGCTCAAATGGATGGGCT TGGCGATTGGTTCTTCTGGACATGGAAGATTGGCGAATCAGAGCGCGGTATTGTTGAATCGCCCTTCTGGTCCTA CCAGCTTGGTCTCCGAAATGGATGGATGCCGACCGACCCTCGCACTGTAATTGGTACTTGCCAGGCCCTTGGTGT CTATACCCCATGGGATGGAACCTTCCAACCATACATGATTGGAGGTCCTGGTGCAGGGGCACCTTCCGATGCTTC CAGCTATCCATGGCCACCCACCGCGCTCGGTGATGGCAATGCTGTCGCAGCACTTCCCACTTATGCTACCACCGG CAGTCCTGTCGTCCTGCCCGCACCCACTTTCACCGATTCGCAAGGCAACGAGATCCTTCCTAGCGGCACTCAGGC AAGCCACCCCGCGCCAACACCCATCGCCGGATGCACCTATCCTGATGGCTGGGACTCTGACGGTGTACCACCCCC TGCGCTTTGTGGAGTGTAATTCCGCAGTTTTTCCCGGGCCCTGGGCCCTCGAGCTATATATGGACTCTAGATACC CTCTTTCCACGACAGCTATTCTTTCGCTACGAACTCCTTTTTTCTGACTTGTGACGCTTTCCGGACGCTTTATTA TATACCTTTAATGCTGATAAAACGCGGACGCTTGACCTTTCTCTGACTTTCCGATACACAACGCTGTACTGACGC CTCGGTATCGTAGCTTTAGAGCCATTGAGACCGCGCTCTCCCCTTTC |
| Length | 2522 |
Gene
| Sequence id | CopciAB_481308.T0 |
|---|---|
| Sequence |
>CopciAB_481308.T0 AGAGTGCACTTGCGGTCTATAATTCACCATGGTTGGCAGGTGGCAAGCGGATTGAGCGCCTGCTTCCCTTTCTTC CCTCTCCCTCCCTCCCTCCTCGACTGGTCCCATGGCAGAGCGACCAGTTTCGATAGTGTCGGACGGCGCATTGGC GAATGCAGCATCGGTCCCTCTCCCTCAAGGAGATGGGAACTCGCTCAACGACCCTCAAGCAGCGCTCGACTCGCG GACGCCCTCCTCTGCTGCCTTGCGCGACTCCGCTGCACTAGCGGATTCAACTACAGCGCTCGGCCCCGGCAATAA TGCAGTGGATTCATCGAAAGAAGCGGCGGATTCAGCAGCGGAAGGTGGCCAGGCTGGTGAGCCTGCCTCCACCTC GAGGAAACGACGACCCCTTCTCATTGCCCTCCTGGCCCTGCTCGCGCTCGCAATTGTGGTGCTGGCCGTTGTCCT CCCGGTCTATTTCAAGGTCATCAAACCCCGACAAAATCGAGATGGCTCGACCGATGGGTCCAGCGGCAGCGGCAC CGACGGTAACGAATCGACATCAGCCCCTTCGCCTACCGGGTCTGCTACTCCCACCAACCCTACGTCTGGTGGTGA CGGATCGACAGTCACGACCGAAGATGGAACTACGTTTACCTATTCCAACCAGTTTGGCGGTTTCTGTACGTTTGA CGTCGCTTCTGAGGTTCTGCTTCTCAAGATCACCTTCGATGTGGTGTTGATTCTGTCCTCGCTTTCTCTCTTGAC TTCATACCGCGGCCGCAACTGACTTTCTTATATAGGGTATTCGGATCCCGACGATCCTTACAATGACAGCGCCCA ACCAAACTCGTGGACACCTCCGCTGAACGCTAGCTTCGATTACACGACGACTCGCATCTTTGGGTGGGTCTTCCT ACGACAAGGAAAGGATAAGAGAGGTCTCTAACTCTTGACAGCGTGAATCTTGGTGGTCTCTTCGTATTGGAACCT TTTATCTCCCCTGCTCTATTCCAGAGATACCCTGGTGCCAGAGATGAATGGGATCTCAGTGTAGCTATGGCCGCA GATACCGCGAACGGCGGTCTCGATCAACTGGAGGAACATTACAGGACATTCATCGTACGTCATCGTTGAGCCTTA CCCCCGAACCACAGCTCATTCTTGTTTTTGCCGAAACAGACGGAACAAGACATCGCTGAGATTGCGGGCGCCGGC TTGAACTGGGTCCGTCTCCCTGTTCCTTTCTGGGCGATAGACAAGTGGCCTGGTGAGCCCTTCCTTGAGAGGACT TCCTGGAGGTGAGGGATAGTTCAATTACAATCAGACACGCAGGTGAATTACTCGCTCTGTTCTTCCAGATATATC GTTCGAGTGCTGCAGTGGTGTCGAAAATACGGTCTCCGTGTGAACCTCGATCTTCACACTATCCCTGGTTCACAC AATGGTATGTCACGTTTTCGCAACCAAGGATCACTGTCTAACCTTGCCCCAAGCTTACAACCACGGCGGAAAACT GAATGCGTTCAATTTCCTCAACGGCGCGATGGGCATGGCAAACGCCCAGAGGGCTTTGTACTATATCCAGGTATT CACCGAGTTCATCAACCAGCCTGAATGGCGAAACGTCGTACCGATGTTCAGCATCATGAATGAGCCTATCATTGG CACCATCGGTGTTGATCAATTGCGTTCGTTGTAAGTCCAACGCACCGTTCAAAATTCGTCTTTCGCTTACCCTAC AAATGTGCTCAGCTACGTCGAAGCGCATCGCATTATGCGCGAAATTACTGGATACGGAGAAGGCAATGGACCATA CATGGTCATCCACGATAGTTTCCGTGGTCCTGGTCCGTGGGCTGGCTTCATGACTGGCGCTGATCGAGTGGGAAT GGACGTTCACCCTTACTTCGCCTTCAATGGTGATTCCGACCCTCCGACTATCGACGGTGGTGTTGGTCCTGGTGC AGGCAACGGTTGGCCTCTTCGCGCATGTAATCGATTCAATGCCATGATGAATGACAGGTATGCCCTGTCACCCTA TCCCTCACCATTCCGATGTTAATCATCCATTACAGTCGAAGGGATTTCGGTGTTACAGTCGCTGGGGAATTCAGC AACGCCTGGCAAGATTGCTCTCTCTTCCTTCGTGGAGTAGGTGGGAGGGCGGACTACGGTGGTGACTGCACCCCC TGGCTGGACTCTTCCGGTTGGTCGGATGGTGTCAAGGCCGGCCTTTTGGCGTTTGCTTCTGCTCAAATGGATGGG CTTGGCGATTGGTTCTTCTGGACATGGAAGGTCTGTACACTTCCAGTTCTGTGGACACTTCCTTAAACTGATAAC TATCCCATAGATTGGCGAATCAGAGCGCGGTATTGTTGAATCGCCCTTCTGGTCCTACCAGCTTGGTCTCCGAAA TGGATGGATGCCGACCGACCCTCGCACTGTAATTGGTACTTGCCAGGCCCTTGGTGTCTATACCCCATGGGATGG AACCTTCCAACCATACATGATTGGAGGTCCTGGTGCAGGGGCACCTTCCGATGCTTCCAGCTATCCATGGCCACC CACCGCGCTCGGTGATGGCAATGCTGTCGCAGCACTTCCCACTTATGCTACCACCGGCAGTCCTGTCGTCCTGCC CGCACCCACTTTCACCGATTCGCAAGGCAACGAGATCCTTCCTAGCGGCACTCAGGCAAGCCACCCCGCGCCAAC ACCCATCGCCGGATGCACCTATCCTGATGGCTGGGACTCTGACGGTGTACCACCCCCTGCGCTTTGTGGAGTGTA ATTCCGCAGTTTTTCCCGGGCCCTGGGCCCTCGAGCTATATATGGACTCTAGATACCCTCTTTCCACGACAGCTA TTCTTTCGCTACGAACTCCTTTTTTCTGACTTGTGACGCTTTCCGGACGCTTTATTATATACCTTTAATGCTGAT AAAACGCGGACGCTTGACCTTTCTCTGACTTTCCGATACACAACGCTGTACTGACGCCTCGGTATCGTAGCTTTA GAGCCATTGAGACCGCGCTCTCCCCTTTC |
| Length | 3029 |