CopciAB_492531
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_492531 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 492531 |
| Uniprot id | Functional description | Belongs to the GMC oxidoreductase family | |
| Location | scaffold_13:517558..521228 | Strand | - |
| Gene length (nt) | 3671 | Transcript length (nt) | 2156 |
| CDS length (nt) | 1953 | Protein length (aa) | 650 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | NCU01853 |
| Aspergillus nidulans | AN0567 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Lentinula edodes NBRC 111202 | Lenedo1_1134174 | 89 | 0 | 1243 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_116682 | 87.6 | 0 | 1225 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1458535 | 86.8 | 0 | 1224 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_195553 | 87.3 | 0 | 1223 |
| Agrocybe aegerita | Agrae_CAA7264498 | 86.4 | 0 | 1215 |
| Schizophyllum commune H4-8 | Schco3_2607677 | 86.9 | 0 | 1213 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21821 | 85.5 | 0 | 1197 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_114505 | 85 | 0 | 1193 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21625 | 84.2 | 0 | 1189 |
| Flammulina velutipes | Flave_chr09AA00051 | 86.2 | 0 | 1185 |
| Pleurotus ostreatus PC9 | PleosPC9_1_129097 | 84.2 | 0 | 1185 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1110004 | 83 | 0 | 1177 |
| Auricularia subglabra | Aurde3_1_1359328 | 67.9 | 8.047E-292 | 916 |
| Grifola frondosa | Grifr_OBZ65421 | 77.7 | 7.392E-197 | 623 |
| Lentinula edodes B17 | Lened_B_1_1_15363 | 52.3 | 1.674E-162 | 523 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_492531.T0 |
| Description | Belongs to the GMC oxidoreductase family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 7 | 313 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 427 | 614 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| K17066 |
EggNOG
| COG category | Description |
|---|---|
| E | Belongs to the GMC oxidoreductase family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_3 |
Transcription factor
| Group |
|---|
| No records |
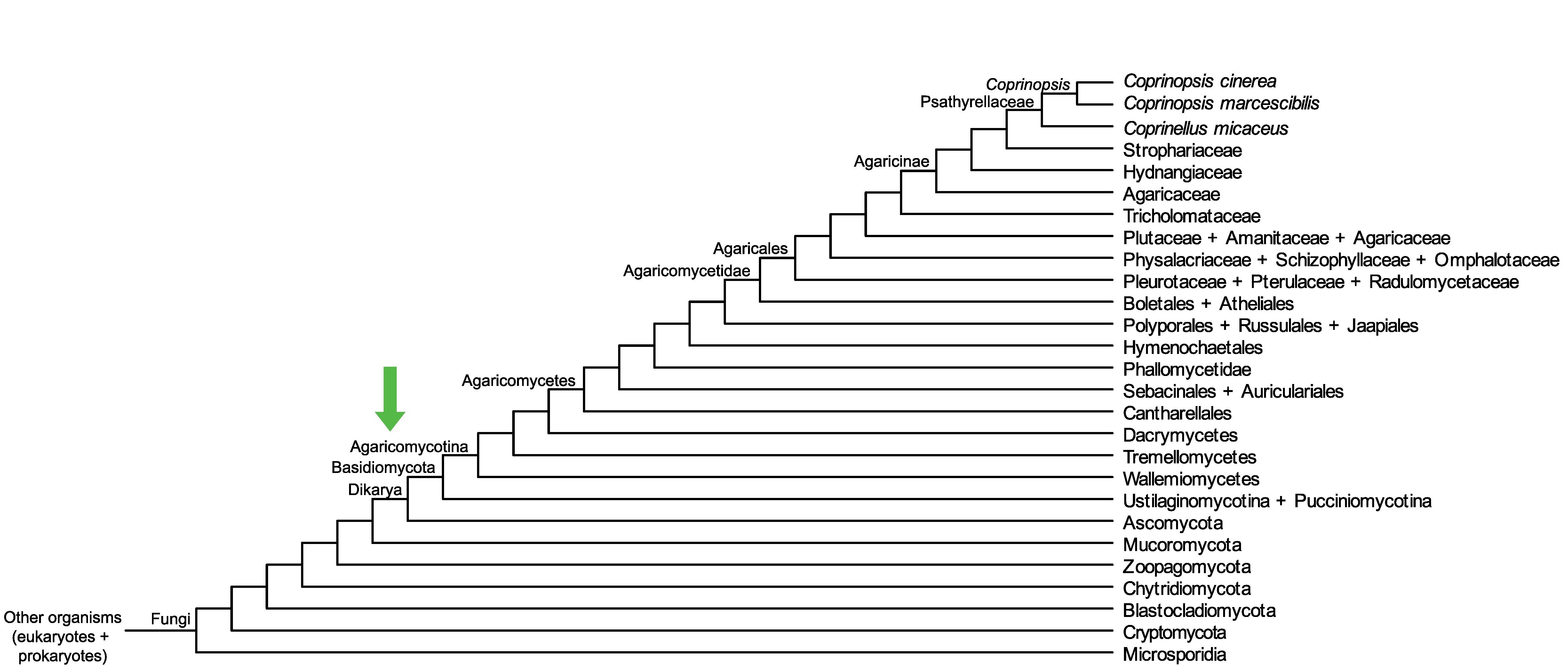
Conservation of CopciAB_492531 across fungi.
Arrow shows the origin of gene family containing CopciAB_492531.
Protein
| Sequence id | CopciAB_492531.T0 |
|---|---|
| Sequence |
>CopciAB_492531.T0 MGHPEEVDIIVAGGGPAGCVVAGRLAYADPNLKVMLIEAGANNRDDPWVYRPGIYVRNMQQNGVNDKATFYTDTK ESSYLRGRRSIVPCANILGGGSSINFQMYTRASASDWDDFKTPGWTAKDLLPLMKRLENYQKPTNNDRHGYDGPI AISNGGTITALAQDFIRASDALGIPFSDDIQDLDTSHASEIWAKYINRHTGRRSDAATAYVHSVMDVQSNLYLRT NAKVNRVIFEGNKAVGVAYVPARNRANGGKLLETIVKARKMVVLSSGTLGTPQILERSGVGAAPLLKQLDIKVVS DLPGVGEEYQDHYTTLSLYRVSNESVTADDFLRGDPEVQKELFAEWETSPEKARLSSNFIDAGFKIRPTEEELKE MGPEFNALWNRYFKDKPDKPVMFGSIVSGAYADHTLLPPGKYITMFQYLEYPASRGKIHIKSPNPYVEPFFDSGF MNNKADFAPIRWSYKKTREVARRMDAFRGELTSHHPRFHPASPAACKDIDIQTAKEILPNGLTVGIHMGTWHKPG DQYDAKKVHDEIKYSKEDDQAIDDWIADHVETTWHSLGTCAMKPREEGGVVDGRLNVFGTQNLKVVDLSICPDNL GTNTYSSALLVGEKGADLIAEDLGLKIRTPHAPVPHAPIPTGKPTTQLVR |
| Length | 650 |
Coding
| Sequence id | CopciAB_492531.T0 |
|---|---|
| Sequence |
>CopciAB_492531.T0 ATGGGTCATCCTGAAGAAGTCGATATCATTGTTGCAGGTGGAGGCCCAGCCGGCTGTGTTGTCGCCGGCCGCTTG GCCTACGCCGACCCCAACCTCAAGGTCATGCTGATCGAAGCTGGAGCCAATAATCGTGATGACCCTTGGGTCTAC AGGCCAGGAATCTATGTCCGCAATATGCAGCAGAATGGGGTGAACGACAAGGCTACGTTCTATACTGACACTAAG GAATCGTCGTACCTCCGTGGTCGAAGGAGCATTGTTCCATGTGCCAACATTCTGGGTGGAGGCAGTAGCATTAAT TTCCAGATGTACACCAGGGCTTCTGCTTCCGATTGGGACGACTTCAAAACTCCGGGATGGACTGCGAAAGACCTC CTTCCTTTGATGAAGCGTCTCGAGAATTATCAGAAGCCGACTAACAACGATAGGCATGGATATGATGGACCTATT GCTATCAGTAATGGTGGAACTATCACTGCCCTCGCGCAGGACTTTATTAGGGCGTCGGATGCTTTGGGTATCCCG TTCAGCGACGATATTCAGGATCTGGATACCAGCCACGCCTCGGAGATCTGGGCTAAGTATATCAATCGTCACACC GGAAGGCGTAGCGATGCAGCCACTGCCTACGTCCACAGCGTCATGGACGTCCAGAGCAACCTCTACCTCCGCACT AACGCCAAAGTCAATCGCGTTATCTTTGAAGGCAACAAGGCTGTGGGCGTTGCATATGTTCCCGCTCGCAACAGG GCCAATGGAGGAAAGCTCCTCGAGACCATCGTCAAGGCGCGCAAGATGGTCGTTCTGAGCAGTGGAACTTTGGGA ACGCCGCAGATTCTTGAGAGGTCTGGTGTTGGAGCTGCGCCGTTGTTGAAGCAGTTGGATATTAAGGTTGTTAGT GATCTTCCTGGTGTTGGAGAGGAATATCAAGACCACTACACCACGCTCAGTCTCTACCGCGTCTCGAATGAGAGT GTCACTGCCGATGACTTCCTTCGTGGTGATCCAGAGGTTCAGAAGGAGCTGTTCGCCGAATGGGAGACGAGCCCA GAGAAAGCCAGACTTTCATCCAACTTCATCGATGCAGGCTTCAAGATCCGTCCTACTGAGGAAGAGCTCAAGGAA ATGGGTCCCGAATTCAACGCACTCTGGAACAGGTATTTCAAGGATAAGCCGGACAAGCCTGTGATGTTTGGATCC ATCGTCTCGGGTGCTTATGCCGATCATACCTTGCTTCCTCCCGGGAAGTATATTACCATGTTCCAATACCTCGAG TATCCCGCTAGTCGTGGAAAGATTCACATCAAATCCCCAAACCCCTACGTCGAGCCCTTCTTCGACAGCGGCTTC ATGAACAACAAGGCCGACTTTGCGCCTATCCGATGGAGCTACAAGAAGACTCGTGAAGTCGCTCGGCGGATGGAC GCCTTCCGCGGTGAATTGACCTCCCACCATCCTCGCTTCCACCCTGCTTCTCCAGCGGCTTGCAAGGACATCGAT ATCCAGACTGCGAAGGAGATTCTTCCCAACGGCTTGACTGTGGGTATTCATATGGGTACTTGGCACAAACCCGGC GATCAGTACGACGCTAAGAAGGTCCATGATGAGATCAAGTACAGCAAGGAAGATGACCAGGCCATCGACGACTGG ATTGCTGACCACGTCGAGACTACATGGCACAGTTTGGGTACTTGCGCCATGAAACCTCGTGAGGAAGGTGGTGTC GTTGACGGTCGTCTCAACGTCTTCGGTACGCAGAACTTGAAGGTTGTCGATTTGAGTATCTGCCCGGACAACCTT GGAACCAACACGTACTCCTCCGCTCTCTTGGTTGGCGAGAAGGGTGCAGACCTTATCGCTGAGGACTTGGGCCTC AAGATCAGGACTCCCCATGCTCCTGTTCCTCATGCCCCCATTCCTACCGGCAAACCCACCACCCAACTTGTCCGC TGA |
| Length | 1953 |
Transcript
| Sequence id | CopciAB_492531.T0 |
|---|---|
| Sequence |
>CopciAB_492531.T0 AGAGCTAGTTCTAGTTCTATCCTCTCCTAGTATCTTAACAAATAGTCGCCTTACGCCATGGGTCATCCTGAAGAA GTCGATATCATTGTTGCAGGTGGAGGCCCAGCCGGCTGTGTTGTCGCCGGCCGCTTGGCCTACGCCGACCCCAAC CTCAAGGTCATGCTGATCGAAGCTGGAGCCAATAATCGTGATGACCCTTGGGTCTACAGGCCAGGAATCTATGTC CGCAATATGCAGCAGAATGGGGTGAACGACAAGGCTACGTTCTATACTGACACTAAGGAATCGTCGTACCTCCGT GGTCGAAGGAGCATTGTTCCATGTGCCAACATTCTGGGTGGAGGCAGTAGCATTAATTTCCAGATGTACACCAGG GCTTCTGCTTCCGATTGGGACGACTTCAAAACTCCGGGATGGACTGCGAAAGACCTCCTTCCTTTGATGAAGCGT CTCGAGAATTATCAGAAGCCGACTAACAACGATAGGCATGGATATGATGGACCTATTGCTATCAGTAATGGTGGA ACTATCACTGCCCTCGCGCAGGACTTTATTAGGGCGTCGGATGCTTTGGGTATCCCGTTCAGCGACGATATTCAG GATCTGGATACCAGCCACGCCTCGGAGATCTGGGCTAAGTATATCAATCGTCACACCGGAAGGCGTAGCGATGCA GCCACTGCCTACGTCCACAGCGTCATGGACGTCCAGAGCAACCTCTACCTCCGCACTAACGCCAAAGTCAATCGC GTTATCTTTGAAGGCAACAAGGCTGTGGGCGTTGCATATGTTCCCGCTCGCAACAGGGCCAATGGAGGAAAGCTC CTCGAGACCATCGTCAAGGCGCGCAAGATGGTCGTTCTGAGCAGTGGAACTTTGGGAACGCCGCAGATTCTTGAG AGGTCTGGTGTTGGAGCTGCGCCGTTGTTGAAGCAGTTGGATATTAAGGTTGTTAGTGATCTTCCTGGTGTTGGA GAGGAATATCAAGACCACTACACCACGCTCAGTCTCTACCGCGTCTCGAATGAGAGTGTCACTGCCGATGACTTC CTTCGTGGTGATCCAGAGGTTCAGAAGGAGCTGTTCGCCGAATGGGAGACGAGCCCAGAGAAAGCCAGACTTTCA TCCAACTTCATCGATGCAGGCTTCAAGATCCGTCCTACTGAGGAAGAGCTCAAGGAAATGGGTCCCGAATTCAAC GCACTCTGGAACAGGTATTTCAAGGATAAGCCGGACAAGCCTGTGATGTTTGGATCCATCGTCTCGGGTGCTTAT GCCGATCATACCTTGCTTCCTCCCGGGAAGTATATTACCATGTTCCAATACCTCGAGTATCCCGCTAGTCGTGGA AAGATTCACATCAAATCCCCAAACCCCTACGTCGAGCCCTTCTTCGACAGCGGCTTCATGAACAACAAGGCCGAC TTTGCGCCTATCCGATGGAGCTACAAGAAGACTCGTGAAGTCGCTCGGCGGATGGACGCCTTCCGCGGTGAATTG ACCTCCCACCATCCTCGCTTCCACCCTGCTTCTCCAGCGGCTTGCAAGGACATCGATATCCAGACTGCGAAGGAG ATTCTTCCCAACGGCTTGACTGTGGGTATTCATATGGGTACTTGGCACAAACCCGGCGATCAGTACGACGCTAAG AAGGTCCATGATGAGATCAAGTACAGCAAGGAAGATGACCAGGCCATCGACGACTGGATTGCTGACCACGTCGAG ACTACATGGCACAGTTTGGGTACTTGCGCCATGAAACCTCGTGAGGAAGGTGGTGTCGTTGACGGTCGTCTCAAC GTCTTCGGTACGCAGAACTTGAAGGTTGTCGATTTGAGTATCTGCCCGGACAACCTTGGAACCAACACGTACTCC TCCGCTCTCTTGGTTGGCGAGAAGGGTGCAGACCTTATCGCTGAGGACTTGGGCCTCAAGATCAGGACTCCCCAT GCTCCTGTTCCTCATGCCCCCATTCCTACCGGCAAACCCACCACCCAACTTGTCCGCTGAAGCCGCCAATCAGTC ATATCTTGAAGCCTTCATACCATGTGTAGCGAATGGTGCAGTGCGCCGATCTTGGGTATCATGATGTCGTGAAGA TTGAGAAGAAGAAATCATTGTATCATTATATGTTGAATAAACCTGTCGTTGATTAC |
| Length | 2156 |
Gene
| Sequence id | CopciAB_492531.T0 |
|---|---|
| Sequence |
>CopciAB_492531.T0 AGAGCTAGTTCTAGTTCTATCCTCTCCTAGTATCTTAACAAATAGTCGCCTTACGCCATGGGTCATCCTGAAGAA GTCGATATCATTGTTGCAGGTGGAGGCCCAGCCGGTAGGGATGCCTCATCTGTACGACTATGAAAAACACTAACG GAATCTCAGGCTGTGTTGTCGCCGGCCGCTTGGCCTACGCCGACCCCAACCTCAAGGTCATGCTGATCGAAGGTT CGCATTGCTCTCTGATTGTTCAGTCAGAAGTGAGGCTGAATATACTTGGCTCCAGCTGGAGCCAATAATCGTGAT GACCCTTGGGTCTACAGGTCGGTCCGTCCCTGCTCGACCATTCCAGCGACGTTGACTTGTTTCTCCAAAAGGCCA GGAATCTATGTCCGCAATATGCAGCAGAATGGGGTGTAAGCTGGTATCTTCAAGAGAAGGCAGATAGACGACTGA CCAGTTCTTTCAGGAACGACAAGGCTACGTTCTATACTGACACTAAGGAATCGTCGTACCTCCGTGGTCGAAGGA GCATTGTTCCGTGCGTTATCCGCCTTCGTCGACTCTGAGGGTGCTTGACTGACGGCTTTTCAGATGTGCCAACAT TCTGGGTATGTCTTTACCTCCTCATTGAATATCACACTAAAACCAGCCCTGATAAAGGTGGAGGCAGTAGGTAAG CGTGTTTATGTGTAGTCCGTCCTACCCGACGATTGACCCCACCCTTCTCCACAGCATTAATTTCCAGGTATGTTC CCTTATATCCCATTCTCCTCCAGAAGACTAATGCGATGAGGCACACAGATGTACACCAGGGTAAGCCATGCGTCC CTTGTTCCCCCTCTTCATCCACTTATCTTGACGCCAGGCTTCTGCTTCCGATTGGGGTAAGTCACATCACTTCCG CGGCGTTGTCAATTTGGGAATGACCCTTTCCTTCTTTAGACGACTTCAAAACTCCGGGATGGACTGCGAAAGGTC GGTGTACGGTGCTTGCATTTTCGCTCTCATATGACTGAAGGGTGTTATGGTGCTAGACCTCCTTCCTTTGATGAA GCGTGTACGCCGTTCGAATATGTTTCTCGATGTTGTATTGATTTCTTGCTAGCTCGAGAATTATCAGAAGCCGAC TAACAACGATAGGCATGGATATGATGGACCTATTGCTATCAGTAATGGTGGAACTATCACTGCCCTCGCGCAGGA CTTTATTAGGGCGTCGGATGCTTTGGGTATCCCGTTCAGCGGTGCGTCCCTTTCCGTAATTTTACTCGCCTTATC TCACACGTTTCAGACGATATTCAGGATCTGGATACCAGCCACGCCTCGGAGATCTGGGCTAAGTATATCAATCGT CACACCGGAAGGCGTAGCGATGCAGCCACTGGCGAGTCATACTATCACCCATCCTACACACGTGCTGACCCAAGC CATAGCCTACGTCCACAGCGTCATGGACGTCCAGAGCAACCTCTACCTCCGCACTAACGCCAAAGTCAATCGCGT TATCTTTGAAGGCAACAAGGCTGTGGGCGTTGCATATGTTCCCGCTCGCAACAGGGCCAATGGAGGAAAGCTCCT CGAGACCATCGTCAAGGCGCGCAAGATGGTCGTTCTGAGCAGTGGAACTTTGGGAACGCCGCAGATTCTTGAGAG GTCTGGTGTTGGAGCTGCGCCGTTGTTGAAGCAGTTGGATATTAAGGTTGTTAGTGATCTTCCTGGTGTTGGGTA TGTTTTGAGCTTGAAAGGATTGTGGGTGTGATGTTGACTTGTTATATCTATTACAGAGAGGAATATCAAGACCAC TACACCACGCTCAGTCGTATGTCCTCGGCTGCGATTTTTGGTCAGTGACAATAGGCTGATGTCATCGACAGTCTA CCGCGTCTCGAATGAGAGTGTCACTGCCGATGACTTCCTTCGTGGTGATCCAGAGGTTCAGAAGGAGGTAGGGCT CCGAGCCTTAACGCGAACTCGACTGACTGGAGTACCCTATAGCTGTTCGCCGAATGGGAGACGAGCCCAGAGAAA GCCAGACTTTCATCCAACTTCATCGATGCAGGCTTCAAGGTACGAGGGTTGCGTCTCTCTAGTATTTCGTTTGGT CGCTGAGGAAGTGGAATGACTTCTGCTAGATCCGTCCTACTGAGGAAGAGCTCAAGGAAATGGGTCCCGAATTCA ACGCACTCTGGAACAGGTATTTCAAGGATAAGCCGGACAAGCCTGTGATGTTTGGATCCATCGTCTCGGGTGGTA TGTGTTACGCTGCTCTAGGAAATCATTCGTTGCTCACTACCCCACACTCCGATAATAGCTTATGCCGATCATACC TTGCTTCCTCCCGGGAAGTATATTACCATGTGAGTTCTTATCCGCGATTGCTAGCCAGGACATTTCTGACTGTGC CCCACGATATGAAGGTTCCAATACCTCGAGTATCCCGCTAGTCGTGGAAAGGTAGGCTGCTGCTCCAATATACCA GCCACCCCCTTCCCCTGAACATACGTCTCGGCCAGATTCACATCAAATCCCCAAACCCCTACGTCGAGCCCTTCT TCGACAGCGGCTTCATGAACAACAAGGCCGACTTTGCGCCTATCCGATGGAGCTACAAGAAGACTCGTGAAGTCG CTCGGCGGATGGACGGTACGCTGCCACGCCTTGACCTGGAATAGGACAGCCGTCGCCTCTGACTGGGTGTATTGT CTAGCCTTCCGCGGTGAATTGACCTCCCACCATCCTCGCTTCCACCCTGCTTCTCCAGCGGCTTGCAAGGACATC GATATCCAGACTGCGAAGGAGATTCTTCCCAACGGCTTGACTGTGGGTATTCATATGGGTACTTGGCACAAGTAC GTGATGGTGACCCTATGAAAGAGAGCGGGAGCTGACCCAACTTGCCAATGTAGACCCGGCGATCAGTACGACGCT AAGAAGGTCCATGATGAGATCAAGTACAGCAAGGAAGATGACCAGGCCATCGACGACTGGATTGCTGACCACGTC GAGACTACATGGCACAGTTTGGGTGCGTACTTCCCATGGCTGAATCATGATCATATTGCTGGATGCTGATCCTTG TTCAGGTACTTGCGGTAAGGATTCTCCCTCCCCACGCGCTGCACAAACAGGGATACTGACAATTGCGATGATGCG TAGCCATGAAACCTCGTGAGGAAGGTGGTGTCGTTGACGGTCGTCTCAACGTCTTCGGTACGCAGAACTTGAAGG TTGTCGATTTGAGTATCTGCCCGGTAGGCCATTTCCTGAAAGGACAGCCAGCGAATTGGAGAGTTGACTTTGTCT GCAGGACAACCTTGGAACCAACACGTACTCCTCCGCTCTCTTGGTTGGCGAGAAGGGTGCAGACCTTATCGCTGA GGACTTGGGTGAGTACCTTCTCGTGACGCGAAGATTTGCTGAGGCTGACGGCGATCGAACGTGACAGGCCTCAAG ATCAGGACTCCCCATGCTCCTGTTCCTCATGCCCCCATTCCTACCGGCAAACCCACCACCCAACTTGTCCGCTGA AGCCGCCAATCAGTCATATCTTGAAGCCTTCATACCATGTGTAGCGAATGGTGCAGTGCGCCGATCTTGGGTATC ATGATGTCGTGAAGATTGAGAAGAAGAAATCATTGTATCATTATATGTTGAATAAACCTGTCGTTGATTAC |
| Length | 3671 |