CopciAB_492701
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_492701 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | HRK1 | Synonyms | 492701 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_2:1839582..1843187 | Strand | - |
| Gene length (nt) | 3606 | Transcript length (nt) | 3463 |
| CDS length (nt) | 2718 | Protein length (aa) | 905 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7271782 | 53.6 | 8.14E-277 | 868 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1060875 | 55 | 2.599E-258 | 814 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_68677 | 52.3 | 3.171E-243 | 770 |
| Pleurotus ostreatus PC9 | PleosPC9_1_102642 | 52 | 7.611E-243 | 769 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_199602 | 53.4 | 1.364E-232 | 739 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_19776 | 45.1 | 1.022E-229 | 731 |
| Lentinula edodes NBRC 111202 | Lenedo1_902075 | 46.7 | 1.104E-228 | 728 |
| Schizophyllum commune H4-8 | Schco3_2485669 | 51 | 5.451E-218 | 697 |
| Grifola frondosa | Grifr_OBZ75840 | 45.7 | 2.152E-192 | 622 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1380915 | 66.5 | 9.331E-167 | 547 |
| Lentinula edodes B17 | Lened_B_1_1_247 | 43.6 | 1.245E-133 | 449 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_20085 | 88.6 | 1.944E-119 | 407 |
| Auricularia subglabra | Aurde3_1_33220 | 86.7 | 3.475E-118 | 404 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | HRK1 |
|---|---|
| Protein id | CopciAB_492701.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 261 | 464 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| D | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
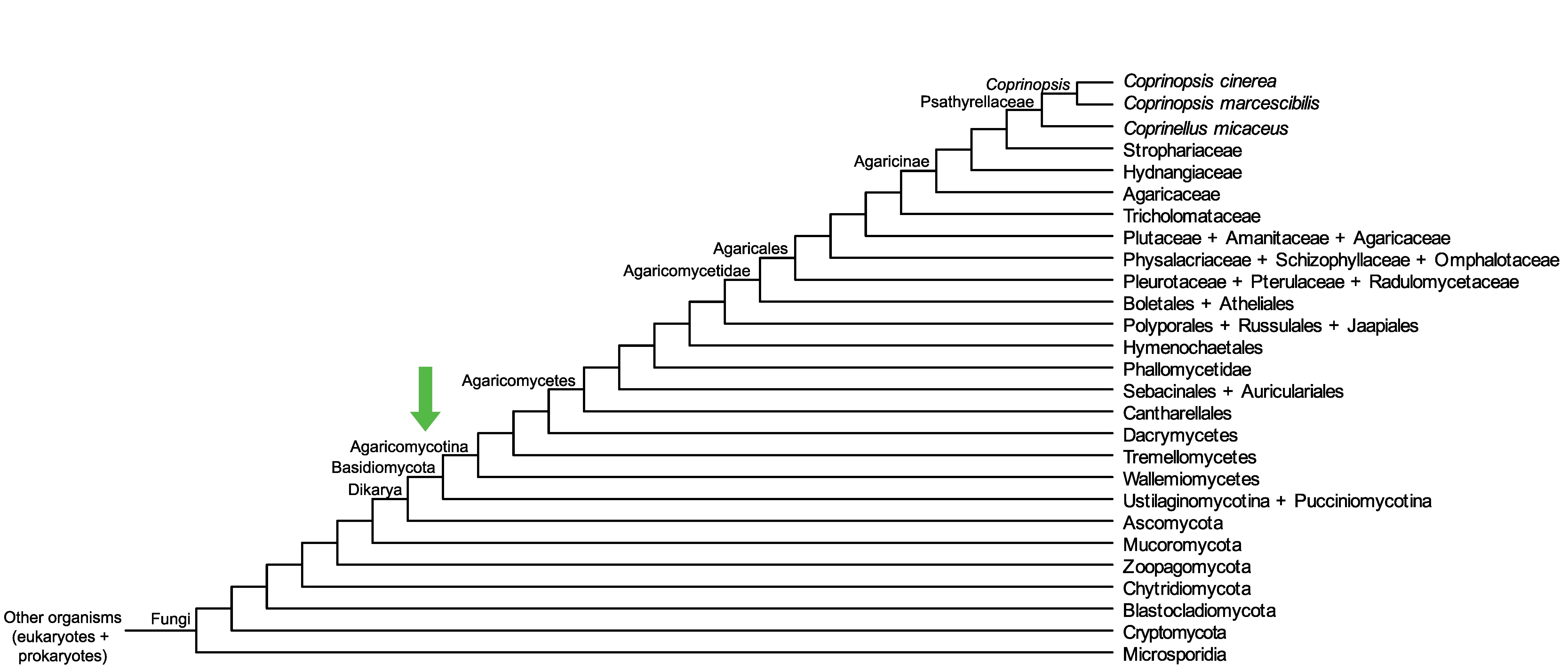
Conservation of CopciAB_492701 across fungi.
Arrow shows the origin of gene family containing CopciAB_492701.
Protein
| Sequence id | CopciAB_492701.T0 |
|---|---|
| Sequence |
>CopciAB_492701.T0 MSSDVVQPTTVSDGAKASFPLTLAVPDDEQPGPVFRRSKSPSQAAIEQSSQQHKSTLEKPEKPERSSRFSFIRKQ SFSLLMPEPSTPSTPKHGPLHDLKRFLNNHIPHSHHHHNQSRSSTQAASTPRRSSVGSSSGPLSPAGARESPTPL DGRPEPKILADVRNSKKKTRSPSPIRQIKDRTPSLKSLRSLKGSSRSSQPPSPTSPQHKTEGSKSKHHHHHHHHR HQSPNRRATSRSHHPITSLHDATQAHLSKKYGKWGRVLGSGAGGTVRLIKGNAKTGGAIYAVKEFRPKRSGETEK EYQKKVTAEFCVGSTLKHPNIIETIDIVQDRGHYYEVMEYAPYDLFSVVMSGKMCRPEIYCVFRQICDGVEYLHE MGLAHRDLKLDNCVMTADNVVKLIDFGTATVFHYPGKAHTPATGIVGSDPYLAPEVLQQDSYDPRKTDVWSVAII FLCMVLRRFPWKIPDPKTDPSFKAFVNAHPDLSAKPVPKPKKLTAEKNVTDQEGASRNLAISTPGRQMTVPPSAH SSGPMDRMPPRERASSTDISIGPESSSTSNSSETTSIRTHTSAGTESTEVTDPPSRFASGSAGHSLNTITDDAKA AKDLSPADARRKSFLQSRGYPHPPTSRSAMTLPLTFTGSPAVRRDSDSSQEMDPSVLTFARPGNSTESLPLSPYL LQDVPTPKGNSLPRAILPARAGSDTVLPAGGIAPGEPLSSEAPPIIVQEPEEETPTPSAAAPAAIKEESTPVVAG PPPPPDSTVPKPRRRQRAESVTTFHGGGAETIFRLLPRETRPALRRMLYVEPASRCTLTDLLKGRGKTSDLLCGC DLAKSKEERLAEMESYRHRHGTDSPTQVSCVDHDECDSEDDDDGDEWLTSIVPCSRPGVQPQHVHIKVQVDEKAG KRRFF |
| Length | 905 |
Coding
| Sequence id | CopciAB_492701.T0 |
|---|---|
| Sequence |
>CopciAB_492701.T0 ATGTCCAGCGATGTTGTGCAGCCCACCACCGTTTCCGACGGCGCCAAGGCCAGTTTTCCGTTGACGCTCGCTGTC CCGGACGATGAGCAGCCCGGACCAGTATTTCGTCGCAGCAAGTCTCCATCTCAGGCAGCTATTGAACAGTCATCA CAACAACACAAGTCGACATTGGAAAAGCCTGAAAAGCCAGAGAGATCCTCAAGATTCAGCTTTATTCGCAAACAG AGCTTTTCGCTACTGATGCCTGAACCGAGCACTCCCTCGACGCCAAAACATGGCCCTCTCCATGACCTCAAGCGC TTCCTAAACAACCACATACCCCATTCTCATCACCACCACAACCAGTCTCGGTCTTCGACACAGGCTGCCTCTACC CCCCGCAGGTCCTCCGTGGGCTCCAGCAGCGGCCCCTTGTCTCCAGCCGGCGCCCGCGAGTCTCCAACCCCTCTC GACGGCAGGCCAGAACCAAAAATACTAGCGGACGTCAGGAACTCGAAGAAGAAGACACGGAGTCCGTCGCCCATC CGCCAGATCAAGGATAGGACGCCATCGCTCAAGTCGCTAAGGTCGCTCAAGGGCAGCTCTCGCTCATCGCAGCCT CCGTCTCCAACTTCCCCGCAACACAAGACCGAAGGGTCAAAGAGCAAGCACCATCATCATCATCACCACCATCGC CACCAGTCGCCCAATCGTCGTGCAACCTCGCGCTCGCATCATCCCATAACATCGCTGCATGACGCGACCCAGGCG CACCTTTCGAAAAAGTACGGCAAGTGGGGGCGGGTACTTGGCAGCGGCGCCGGTGGCACCGTACGACTTATCAAG GGGAATGCAAAGACTGGAGGAGCGATATATGCTGTGAAGGAGTTTCGTCCAAAGCGCTCGGGAGAGACGGAGAAG GAGTACCAGAAGAAGGTCACAGCCGAGTTTTGCGTGGGCTCCACGTTGAAACACCCCAACATCATCGAGACCATC GACATCGTCCAAGACCGTGGTCACTACTATGAGGTCATGGAGTACGCTCCGTACGACCTCTTTTCGGTGGTCATG TCGGGCAAGATGTGTCGCCCCGAGATCTACTGCGTCTTCCGACAAATCTGCGATGGCGTTGAATACCTTCACGAG ATGGGCTTGGCGCACCGGGACCTCAAACTCGATAATTGCGTCATGACGGCCGACAATGTCGTGAAGCTCATCGAC TTTGGGACAGCGACTGTCTTTCATTATCCTGGAAAGGCCCATACACCAGCCACTGGGATTGTCGGCAGTGATCCC TATCTTGCTCCAGAGGTACTCCAACAGGACTCCTATGACCCGAGGAAGACGGATGTGTGGAGTGTAGCAATCATC TTTCTCTGCATGGTCCTGCGTCGGTTCCCTTGGAAAATCCCCGATCCGAAGACGGACCCAAGCTTCAAGGCTTTC GTCAACGCGCACCCCGACTTGTCCGCAAAGCCTGTGCCAAAGCCTAAGAAGCTTACCGCCGAGAAGAATGTAACT GACCAGGAGGGCGCATCGAGAAACTTGGCCATTTCTACTCCTGGCCGGCAAATGACGGTTCCTCCTTCTGCCCAT TCGAGCGGACCCATGGATAGGATGCCCCCACGGGAGCGCGCATCCAGCACCGACATTTCGATTGGCCCCGAGTCC TCCAGCACCAGCAACTCCTCAGAGACGACCTCAATTCGGACGCATACCTCGGCCGGCACCGAAAGCACAGAAGTG ACTGACCCACCTTCACGTTTCGCCAGTGGGTCCGCCGGGCACAGCTTGAACACAATCACCGATGACGCGAAGGCC GCCAAGGATTTGTCACCAGCAGACGCTCGAAGGAAGAGTTTCCTCCAGTCGAGAGGATACCCGCATCCACCTACA TCCCGGAGTGCGATGACTCTCCCGCTGACCTTCACTGGCTCACCAGCGGTTCGGCGAGATTCGGATTCGTCACAA GAGATGGATCCCTCCGTCTTGACTTTTGCCCGGCCCGGGAATTCAACAGAGTCGCTACCTTTAAGCCCTTATCTC CTCCAAGATGTTCCAACGCCGAAGGGCAACAGTCTGCCTCGGGCTATCCTTCCTGCCCGAGCAGGCTCGGATACG GTCCTACCAGCTGGTGGCATCGCCCCAGGGGAGCCCTTGTCGTCCGAGGCACCTCCTATTATTGTCCAAGAGCCT GAAGAAGAGACACCAACACCATCGGCGGCAGCGCCCGCTGCTATCAAAGAGGAGTCTACGCCGGTTGTTGCTGGA CCACCACCGCCTCCTGACTCAACAGTACCCAAACCTCGTCGACGCCAACGAGCAGAGAGTGTTACGACGTTCCAC GGTGGAGGAGCAGAGACCATCTTTCGGTTACTCCCCCGCGAAACTCGGCCCGCGCTCCGGCGGATGTTGTATGTG GAACCTGCCTCTCGTTGCACCCTCACCGACCTGCTCAAGGGAAGGGGAAAGACCAGCGACCTCCTGTGCGGATGC GACTTGGCCAAGTCAAAGGAAGAGCGGTTAGCGGAGATGGAATCTTACAGGCATCGCCATGGCACAGATTCCCCA ACTCAAGTGTCTTGCGTGGACCACGATGAATGTGATTCAGAGGACGACGACGACGGTGACGAGTGGTTAACGAGC ATCGTTCCATGTTCCAGACCTGGAGTCCAACCCCAGCACGTACACATCAAGGTTCAGGTGGACGAAAAGGCCGGC AAGAGACGATTCTTCTAG |
| Length | 2718 |
Transcript
| Sequence id | CopciAB_492701.T0 |
|---|---|
| Sequence |
>CopciAB_492701.T0 GCTTTCCACCACCATCGCCCACCATGTCCAGCGATGTTGTGCAGCCCACCACCGTTTCCGACGGCGCCAAGGCCA GTTTTCCGTTGACGCTCGCTGTCCCGGACGATGAGCAGCCCGGACCAGTATTTCGTCGCAGCAAGTCTCCATCTC AGGCAGCTATTGAACAGTCATCACAACAACACAAGTCGACATTGGAAAAGCCTGAAAAGCCAGAGAGATCCTCAA GATTCAGCTTTATTCGCAAACAGAGCTTTTCGCTACTGATGCCTGAACCGAGCACTCCCTCGACGCCAAAACATG GCCCTCTCCATGACCTCAAGCGCTTCCTAAACAACCACATACCCCATTCTCATCACCACCACAACCAGTCTCGGT CTTCGACACAGGCTGCCTCTACCCCCCGCAGGTCCTCCGTGGGCTCCAGCAGCGGCCCCTTGTCTCCAGCCGGCG CCCGCGAGTCTCCAACCCCTCTCGACGGCAGGCCAGAACCAAAAATACTAGCGGACGTCAGGAACTCGAAGAAGA AGACACGGAGTCCGTCGCCCATCCGCCAGATCAAGGATAGGACGCCATCGCTCAAGTCGCTAAGGTCGCTCAAGG GCAGCTCTCGCTCATCGCAGCCTCCGTCTCCAACTTCCCCGCAACACAAGACCGAAGGGTCAAAGAGCAAGCACC ATCATCATCATCACCACCATCGCCACCAGTCGCCCAATCGTCGTGCAACCTCGCGCTCGCATCATCCCATAACAT CGCTGCATGACGCGACCCAGGCGCACCTTTCGAAAAAGTACGGCAAGTGGGGGCGGGTACTTGGCAGCGGCGCCG GTGGCACCGTACGACTTATCAAGGGGAATGCAAAGACTGGAGGAGCGATATATGCTGTGAAGGAGTTTCGTCCAA AGCGCTCGGGAGAGACGGAGAAGGAGTACCAGAAGAAGGTCACAGCCGAGTTTTGCGTGGGCTCCACGTTGAAAC ACCCCAACATCATCGAGACCATCGACATCGTCCAAGACCGTGGTCACTACTATGAGGTCATGGAGTACGCTCCGT ACGACCTCTTTTCGGTGGTCATGTCGGGCAAGATGTGTCGCCCCGAGATCTACTGCGTCTTCCGACAAATCTGCG ATGGCGTTGAATACCTTCACGAGATGGGCTTGGCGCACCGGGACCTCAAACTCGATAATTGCGTCATGACGGCCG ACAATGTCGTGAAGCTCATCGACTTTGGGACAGCGACTGTCTTTCATTATCCTGGAAAGGCCCATACACCAGCCA CTGGGATTGTCGGCAGTGATCCCTATCTTGCTCCAGAGGTACTCCAACAGGACTCCTATGACCCGAGGAAGACGG ATGTGTGGAGTGTAGCAATCATCTTTCTCTGCATGGTCCTGCGTCGGTTCCCTTGGAAAATCCCCGATCCGAAGA CGGACCCAAGCTTCAAGGCTTTCGTCAACGCGCACCCCGACTTGTCCGCAAAGCCTGTGCCAAAGCCTAAGAAGC TTACCGCCGAGAAGAATGTAACTGACCAGGAGGGCGCATCGAGAAACTTGGCCATTTCTACTCCTGGCCGGCAAA TGACGGTTCCTCCTTCTGCCCATTCGAGCGGACCCATGGATAGGATGCCCCCACGGGAGCGCGCATCCAGCACCG ACATTTCGATTGGCCCCGAGTCCTCCAGCACCAGCAACTCCTCAGAGACGACCTCAATTCGGACGCATACCTCGG CCGGCACCGAAAGCACAGAAGTGACTGACCCACCTTCACGTTTCGCCAGTGGGTCCGCCGGGCACAGCTTGAACA CAATCACCGATGACGCGAAGGCCGCCAAGGATTTGTCACCAGCAGACGCTCGAAGGAAGAGTTTCCTCCAGTCGA GAGGATACCCGCATCCACCTACATCCCGGAGTGCGATGACTCTCCCGCTGACCTTCACTGGCTCACCAGCGGTTC GGCGAGATTCGGATTCGTCACAAGAGATGGATCCCTCCGTCTTGACTTTTGCCCGGCCCGGGAATTCAACAGAGT CGCTACCTTTAAGCCCTTATCTCCTCCAAGATGTTCCAACGCCGAAGGGCAACAGTCTGCCTCGGGCTATCCTTC CTGCCCGAGCAGGCTCGGATACGGTCCTACCAGCTGGTGGCATCGCCCCAGGGGAGCCCTTGTCGTCCGAGGCAC CTCCTATTATTGTCCAAGAGCCTGAAGAAGAGACACCAACACCATCGGCGGCAGCGCCCGCTGCTATCAAAGAGG AGTCTACGCCGGTTGTTGCTGGACCACCACCGCCTCCTGACTCAACAGTACCCAAACCTCGTCGACGCCAACGAG CAGAGAGTGTTACGACGTTCCACGGTGGAGGAGCAGAGACCATCTTTCGGTTACTCCCCCGCGAAACTCGGCCCG CGCTCCGGCGGATGTTGTATGTGGAACCTGCCTCTCGTTGCACCCTCACCGACCTGCTCAAGGGAAGGGGAAAGA CCAGCGACCTCCTGTGCGGATGCGACTTGGCCAAGTCAAAGGAAGAGCGGTTAGCGGAGATGGAATCTTACAGGC ATCGCCATGGCACAGATTCCCCAACTCAAGTGTCTTGCGTGGACCACGATGAATGTGATTCAGAGGACGACGACG ACGGTGACGAGTGGTTAACGAGCATCGTTCCATGTTCCAGACCTGGAGTCCAACCCCAGCACGTACACATCAAGG TTCAGGTGGACGAAAAGGCCGGCAAGAGACGATTCTTCTAGGTGGTCCTTGGGGACTTCTCCTAGCTTGGAAGGC ATTCGTGCGGTCTCCATTTCGCTTGTACGTTTCATGCTAGCCTTGCGCCATATCTTTAAGTTTTGCTTATCATTC CTTTTTCATCTGCGCCTCCACCCCATTATCACGGGAATTCATCGCATACTTCTATCTTACACGCTCCCTTTTATT TTTATACATCTTAGAAGGTTTTAGTCGACGGATTCTTTGCTTTCACACCTTTCATTGTGAAGCGTGCTTTCCATT TCTACATTCTACATCCCCCAACACCTCGCCATGGCGTCCTCCGCGATGTAATCCGACTGTGGATTTAACACCTCG AGGCCACTCTCAACAGCATGCGCCAACCTGCTCGCAATGGCACCCTTCACACCAACCCGCGCCATTTAAATCCTT CCCTTTCACTCTCGTTTCCTTCATGCGGACACGTTGATTGCTTTTAAAACCGGATTTTTCTCCCCCTTGTCCTAC TGTCGAAATTTAAGATTAAGCACAGGCACTTCTGCGGTCTGTTGTATATTTATCGATTTATTCCCCGCCATCTTT CGCCATGTAGCAATCCCTGCGTACACCATCGCTTCCCTGCTTTCCTGCGACATACGTACCCTTTGAAGCCATAGC ACCTATTATATCGTTATCATCTACTAGTACTTGCTTATCTTTTCCTCCAGATCCCTGTCCCTGTATGACGATTAT GTGCTTTTTCGTG |
| Length | 3463 |
Gene
| Sequence id | CopciAB_492701.T0 |
|---|---|
| Sequence |
>CopciAB_492701.T0 GCTTTCCACCACCATCGCCCACCATGTCCAGCGATGTTGTGCAGCCCACCACCGTTTCCGACGGCGCCAAGGCCA GTTTTCCGTTGACGCTCGCTGTCCCGGACGATGAGCAGGTGGGTTACCCCAGGTCATGATTACACTGCCACCTTT ATCTAATTATATCTCAAACAGCCCGGACCAGTATTTCGTCGCAGCAAGTCTCCATCTCAGGCAGCTATTGAACAG TCATCACAACAACACAAGTCGACATTGGAAAAGCCTGAAAAGCCAGAGAGATCCTCAAGATTCAGCTTTATTCGC AAACAGAGCTTTTCGCTACTGATGCCTGAACCGAGCACTCCCTCGACGCCAAAACATGGCCCTCTCCATGACCTC AAGCGCTTCCTAAACAACCACATACCCCATTCTCATCACCACCACAACCAGTCTCGGTCTTCGACACAGGCTGCC TCTACCCCCCGCAGGTCCTCCGTGGGCTCCAGCAGCGGCCCCTTGTCTCCAGCCGGCGCCCGCGAGTCTCCAACC CCTCTCGACGGCAGGCCAGAACCAAAAATACTAGCGGACGTCAGGAACTCGAAGAAGAAGACACGGAGTCCGTCG CCCATCCGCCAGATCAAGGATAGGACGCCATCGCTCAAGTCGCTAAGGTCGCTCAAGGGCAGCTCTCGCTCATCG CAGCCTCCGTCTCCAACTTCCCCGCAACACAAGACCGAAGGGTCAAAGAGCAAGCACCATCATCATCATCACCAC CATCGCCACCAGTCGCCCAATCGTCGTGCAACCTCGCGCTCGCATCATCCCATAACATCGCTGCATGACGCGACC CAGGCGCACCTTTCGAAAAAGTACGGCAAGTGGGGGCGGGTACTTGGCAGCGGCGCCGGTGGCACCGTACGACTT ATCAAGGGGAATGCAAAGACTGGAGGAGCGATATATGCTGTGAAGGAGTTTCGTCCAAAGCGCTCGGGAGAGACG GAGAAGGAGTACCAGAAGAAGGTCACAGCCGAGTTTTGCGTGGGCTCCACGTTGAAACACCCCAACATCATCGAG ACCATCGACATCGTCCAAGACCGTGGTCACTACTATGAGGTATGCCTATCGGTACATTCTATCACATAATTCCAA GCCCCTCAACTTCTGTTTTCCCAAACTCTCTTTTGTTTTCTTTCTCCAGGTCATGGAGTACGCTCCGTACGACCT CTTTTCGGTGGTCATGTCGGGCAAGATGTGTCGCCCCGAGATCTACTGCGTCTTCCGACAAATCTGCGATGGCGT TGAATACCTTCACGAGATGGGCTTGGCGCACCGGGACCTCAAACTCGATAATTGCGTCATGACGGCCGACAATGT CGTGAAGCTCATCGACTTTGGGACAGCGACTGTCTTTCATTATCCTGGAAAGGCCCATACACCAGCCACTGGGAT TGTCGGCAGTGATCCCTATCTTGCTCCAGAGGTACTCCAACAGGACTCCTATGACCCGAGGAAGACGGATGTGTG GAGTGTAGCAATCATCTTTCTCTGCATGGTCCTGCGTCGGTTCCCTTGGAAAATCCCCGATCCGAAGACGGACCC AAGCTTCAAGGCTTTCGTCAACGCGCACCCCGACTTGTCCGCAAAGCCTGTGCCAAAGCCTAAGAAGCTTACCGC CGAGAAGAATGTAACTGACCAGGAGGGCGCATCGAGAAACTTGGCCATTTCTACTCCTGGCCGGCAAATGACGGT TCCTCCTTCTGCCCATTCGAGCGGACCCATGGATAGGATGCCCCCACGGGAGCGCGCATCCAGCACCGACATTTC GATTGGCCCCGAGTCCTCCAGCACCAGCAACTCCTCAGAGACGACCTCAATTCGGACGCATACCTCGGCCGGCAC CGAAAGCACAGAAGTGACTGACCCACCTTCACGTTTCGCCAGTGGGTCCGCCGGGCACAGCTTGAACACAATCAC CGATGACGCGAAGGCCGCCAAGGATTTGTCACCAGCAGACGCTCGAAGGAAGAGTTTCCTCCAGTCGAGAGGATA CCCGCATCCACCTACATCCCGGAGTGCGATGACTCTCCCGCTGACCTTCACTGGCTCACCAGCGGTTCGGCGAGA TTCGGATTCGTCACAAGAGATGGATCCCTCCGTCTTGACTTTTGCCCGGCCCGGGAATTCAACAGAGTCGCTACC TTTAAGCCCTTATCTCCTCCAAGATGTTCCAACGCCGAAGGGCAACAGTCTGCCTCGGGCTATCCTTCCTGCCCG AGCAGGCTCGGATACGGTCCTACCAGCTGGTGGCATCGCCCCAGGGGAGCCCTTGTCGTCCGAGGCACCTCCTAT TATTGTCCAAGAGCCTGAAGAAGAGACACCAACACCATCGGCGGCAGCGCCCGCTGCTATCAAAGAGGAGTCTAC GCCGGTTGTTGCTGGACCACCACCGCCTCCTGACTCAACAGTACCCAAACCTCGTCGACGCCAACGAGCAGAGAG TGTTACGACGTTCCACGGTGGAGGAGCAGAGACCATCTTTCGGTTACTCCCCCGCGAAACTCGGCCCGCGCTCCG GCGGATGTTGTATGTGGAACCTGCCTCTCGTTGCACCCTCACCGACCTGCTCAAGGGAAGGGGAAAGACCAGCGA CCTCCTGTGCGGATGCGACTTGGCCAAGTCAAAGGAAGAGCGGTTAGCGGAGATGGAATCTTACAGGCATCGCCA TGGCACAGATTCCCCAACTCAAGTGTCTTGCGTGGACCACGATGAATGTGATTCAGAGGACGACGACGACGGTGA CGAGTGGTTAACGAGCATCGTTCCATGTTCCAGACCTGGAGTCCAACCCCAGCACGTACACATCAAGGTTCAGGT GGACGAAAAGGCCGGCAAGAGACGATTCTTCTAGGTGGTCCTTGGGGACTTCTCCTAGCTTGGAAGGCATTCGTG CGGTCTCCATTTCGCTTGTACGTTTCATGCTAGCCTTGCGCCATATCTTTAAGTTTTGCTTATCATTCCTTTTTC ATCTGCGCCTCCACCCCATTATCACGGGAATTCATCGCATACTTCTATCTTACACGCTCCCTTTTATTTTTATAC ATCTTAGAAGGTTTTAGTCGACGGATTCTTTGCTTTCACACCTTTCATTGTGAAGCGTGCTTTCCATTTCTACAT TCTACATCCCCCAACACCTCGCCATGGCGTCCTCCGCGATGTAATCCGACTGTGGATTTAACACCTCGAGGCCAC TCTCAACAGCATGCGCCAACCTGCTCGCAATGGCACCCTTCACACCAACCCGCGCCATTTAAATCCTTCCCTTTC ACTCTCGTTTCCTTCATGCGGACACGTTGATTGCTTTTAAAACCGGATTTTTCTCCCCCTTGTCCTACTGTCGAA ATTTAAGATTAAGCACAGGCACTTCTGCGGTCTGTTGTATATTTATCGATTTATTCCCCGCCATCTTTCGCCATG TAGCAATCCCTGCGTACACCATCGCTTCCCTGCTTTCCTGCGACATACGTACCCTTTGAAGCCATAGCACCTATT ATATCGTTATCATCTACTAGTACTTGCTTATCTTTTCCTCCAGATCCCTGTCCCTGTATGACGATTATGTGCTTT TTCGTG |
| Length | 3606 |