CopciAB_496994
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_496994 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 496994 |
| Uniprot id | Functional description | peptidyl-serine phosphorylation | |
| Location | scaffold_9:1455860..1459325 | Strand | - |
| Gene length (nt) | 3466 | Transcript length (nt) | 3183 |
| CDS length (nt) | 2274 | Protein length (aa) | 757 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_496994.T0 |
| Description | peptidyl-serine phosphorylation |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 479 | 678 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K08789 |
| K19833 |
EggNOG
| COG category | Description |
|---|---|
| T | peptidyl-serine phosphorylation |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
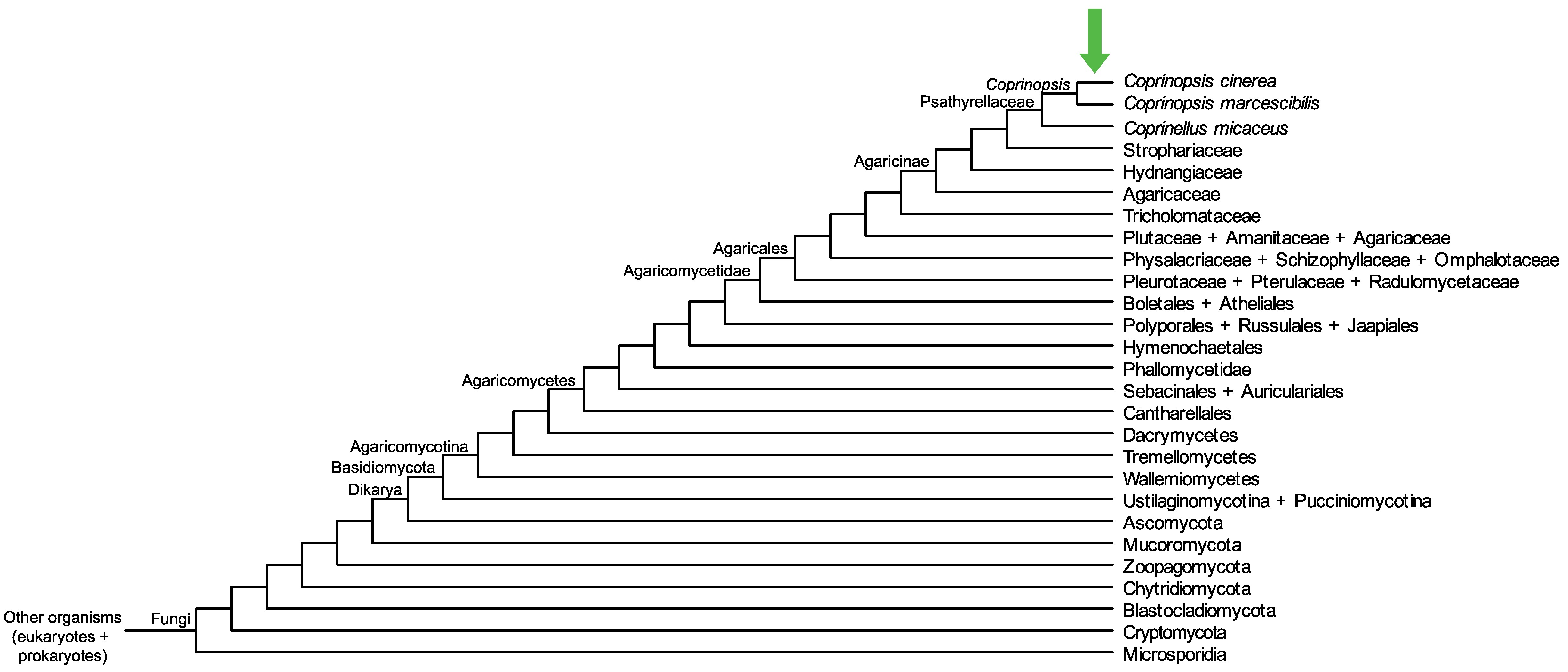
Conservation of CopciAB_496994 across fungi.
Arrow shows the origin of gene family containing CopciAB_496994.
Protein
| Sequence id | CopciAB_496994.T0 |
|---|---|
| Sequence |
>CopciAB_496994.T0 MVRIALWTVMNKLSPSRAKGKSSPRSSPVPSSSPSIHVEELGELSDKENQDPSVVENYQPSPPTVEVIDYTSPPT VEVVDYTSPPTVEVVDYTQTSPPTVEVIDNATSVEEQSSPQPSPVPSSSPSTTTTSVEELSPLSDKESQEPPNAE NHQPRPPSDELDDDLYNVVVIDSPKRPKRSSFILEGFEDDDLYNVCEPKELSPIPGLGLYDVKKDATPTPVPDSD TTAKKDLTPTQEPKSDTTQQEPTPAQEPESDTTQKDPAPTQEEKSDTANQDPTPTQGSESDTTKKDLTPAQKSKS DNKKQVSEGIPGLVYYQKRPLARDDDDTDSPAKKLKANDDSDRTVIPQNNLGFGSRIPTFNFKGSFFRQLELKER IMIVNHQHLGDRRMDTRLFISSVVPATPGDPDTAAIRRSAPRVFVKLLHPSPGYGVNTTEITAYKRIDRLKNILD SEVKKFSIFVLSLEAYISCYGGPTMTTQIFVMPLLKRDLSAALRERNQYDGPSENVQLWIAQIAAGVHFLHNCGI MHRDLKSENILLDAGDNALVTDFGNSFVQVDWINAKLDYDLLYSRANLGTPGWLAPEVEQGLWYGPASDWWGVGA IMLEAMVHPANFYMALSQFRATRLLYPRSRSGADAKFAFETEQTSVPCAFAREVLWMLLHHDPRDRYRYDQLYSH TYFAVYRDGRVVSIFDILEFETAARLNKAKQESGPTSEDPTEDHHQLPYKCDLWRFDHPGTNAEVWLPSYSKIHQ IIDKAYE |
| Length | 757 |
Coding
| Sequence id | CopciAB_496994.T0 |
|---|---|
| Sequence |
>CopciAB_496994.T0 ATGGTACGAATCGCACTGTGGACCGTGATGAACAAACTTTCCCCTTCACGCGCGAAGGGCAAGTCCTCTCCTCGG TCGAGTCCAGTTCCATCCTCCTCCCCTTCGATCCACGTCGAAGAGCTCGGCGAGCTCAGCGACAAGGAGAATCAA GATCCATCTGTCGTCGAGAACTATCAACCATCTCCTCCCACCGTCGAGGTCATCGACTACACATCTCCACCCACC GTCGAGGTCGTCGACTACACATCTCCACCCACCGTCGAGGTCGTCGACTACACTCAAACATCACCTCCCACCGTC GAGGTCATCGATAATGCCACATCCGTCGAAGAACAGTCCTCTCCTCAGCCGAGTCCAGTTCCGTCCTCTTCGCCT TCCACCACCACCACTTCGGTCGAAGAGCTCAGTCCGCTCAGCGACAAGGAGAGTCAAGAGCCACCCAACGCAGAG AACCATCAGCCACGTCCTCCCTCCGACGAGCTCGACGATGATCTTTACAACGTCGTCGTTATCGACTCCCCCAAG CGCCCTAAACGATCTTCTTTCATTCTCGAGGGTTTCGAGGACGACGACTTGTACAACGTTTGCGAGCCGAAGGAA TTGTCCCCCATCCCAGGGCTTGGCTTATACGACGTGAAGAAGGACGCTACACCGACGCCGGTACCAGACTCCGAC ACCACCGCCAAGAAGGATCTTACGCCGACCCAGGAGCCAAAATCCGATACCACCCAGCAGGAGCCTACGCCAGCC CAGGAGCCAGAGTCCGACACCACTCAGAAGGATCCTGCGCCAACCCAAGAGGAAAAGTCCGATACCGCCAACCAG GATCCTACGCCGACTCAGGGGTCAGAGTCAGACACCACCAAAAAGGATCTTACGCCGGCACAGAAGTCGAAATCC GACAACAAGAAGCAAGTATCCGAGGGCATCCCAGGATTGGTCTACTACCAAAAGCGACCACTCGCACGCGACGAC GACGACACTGATTCCCCTGCTAAGAAGCTTAAAGCGAATGACGACTCCGACCGCACTGTTATCCCCCAGAATAAC CTCGGTTTCGGCTCCCGCATTCCCACCTTCAACTTCAAAGGTTCCTTCTTCCGCCAGCTCGAACTCAAAGAGCGC ATCATGATCGTCAACCACCAACACCTTGGAGATCGACGAATGGACACCAGGCTTTTCATCTCGAGTGTTGTCCCC GCGACTCCCGGGGATCCCGACACTGCTGCCATTCGCAGATCAGCACCGCGCGTCTTCGTGAAGCTACTCCATCCA AGCCCCGGATACGGAGTCAACACAACTGAAATTACCGCCTACAAGCGTATCGACAGGCTCAAGAACATACTCGAC TCGGAGGTGAAGAAGTTCAGCATCTTCGTCTTATCCCTGGAAGCATACATATCCTGCTACGGTGGTCCAACTATG ACCACCCAGATCTTCGTCATGCCACTTCTCAAGCGCGATTTGTCTGCTGCGTTGCGAGAGCGAAACCAATACGAC GGACCATCGGAAAACGTTCAACTGTGGATCGCGCAAATTGCGGCCGGGGTCCATTTCCTCCACAATTGTGGTATC ATGCATCGCGATCTCAAGTCAGAGAACATTCTGTTGGATGCAGGCGACAACGCTTTGGTTACCGATTTCGGAAAC TCTTTCGTTCAGGTCGACTGGATCAACGCCAAACTCGATTATGACCTTTTGTACTCTCGTGCCAATTTAGGAACT CCTGGGTGGCTCGCTCCAGAGGTCGAACAAGGTTTGTGGTATGGCCCGGCAAGTGATTGGTGGGGTGTTGGCGCG ATTATGCTCGAGGCCATGGTCCATCCGGCCAACTTCTATATGGCGTTGTCTCAATTCCGGGCCACCCGACTGCTT TACCCGAGGAGCAGGAGCGGGGCAGACGCCAAGTTCGCTTTCGAGACTGAGCAAACCAGCGTCCCTTGCGCGTTT GCGAGAGAAGTCCTCTGGATGTTGCTTCACCACGACCCCCGCGACCGTTACCGTTATGACCAGCTTTACAGCCAC ACTTATTTCGCTGTTTACCGTGATGGCAGAGTGGTATCTATTTTCGACATTTTGGAGTTCGAAACCGCGGCACGA CTGAACAAGGCCAAGCAAGAATCTGGACCAACGTCCGAAGATCCGACTGAGGATCATCATCAGCTTCCCTACAAG TGTGACCTTTGGAGGTTCGACCACCCTGGCACAAATGCGGAAGTGTGGCTCCCATCCTATTCGAAAATCCATCAA ATCATCGACAAGGCTTATGAATAG |
| Length | 2274 |
Transcript
| Sequence id | CopciAB_496994.T0 |
|---|---|
| Sequence |
>CopciAB_496994.T0 GCCGAAGTTTACTGCCAGCGTCGACGCAAGAATTCACCAAAACTCTGGCCATGGGGACGGGGACGTTTCAAAGTC TCATATCGGTGCTCTGGGATGTCTATCCTTTCAAGGTGCTCAACTGGATCTGTATGCTCGGAACTTGCAGTGCTG AACGACGCTCGACGACTTTCTGGACGACGACCCAACTGATTTTTCGGGCGGGTTTATCTGTTTCTCTTTTTCTGG GGGCAGGGACTGCACACAGAGCTCACACCACTGCACTGCTCAACGACCCAGGTGGATGGCGCTCTCGCTGTAGCC CATTGGCACCCAAATCTGTGCCATTCGCAATCGCGATCCCGCCATTGACGAGGACACGGTACGCCGAGAAGCCAC CCAAGCCATCGGAGAACGCCGAGTTTCGTGATCTTGCAACAAAGAGTGTGGGAACCCATTCCAAATGCGGTGGTG ATGTCGATGTCATGGACGCGCGCGTCTGTTGGACCATTGTCCGTTAGGGAGGCTCTCCTTCCAAATCCGTACTTT CGTCCGTCGCGACAACATGACAACTGGCCACTCCTTTGTGAGGGCGCACTCCATTGACATTATCTGCCTCGTGTT CTGGTTAAATGTACATACTTGGAAATTAGAATTCTGCGGACAATGGGCCCGGCCGGCGCTATAAAACAGACGACT CCTGCGATTTCACGGTTCTGCTTTTGCCCACTACCATCGCCTGTCCACCAGTCATATATCGCTCACCACATCCAA CCTTCGTGTCCCCTATACTCTCTTCTCATCTGCCACCATGGTACGAATCGCACTGTGGACCGTGATGAACAAACT TTCCCCTTCACGCGCGAAGGGCAAGTCCTCTCCTCGGTCGAGTCCAGTTCCATCCTCCTCCCCTTCGATCCACGT CGAAGAGCTCGGCGAGCTCAGCGACAAGGAGAATCAAGATCCATCTGTCGTCGAGAACTATCAACCATCTCCTCC CACCGTCGAGGTCATCGACTACACATCTCCACCCACCGTCGAGGTCGTCGACTACACATCTCCACCCACCGTCGA GGTCGTCGACTACACTCAAACATCACCTCCCACCGTCGAGGTCATCGATAATGCCACATCCGTCGAAGAACAGTC CTCTCCTCAGCCGAGTCCAGTTCCGTCCTCTTCGCCTTCCACCACCACCACTTCGGTCGAAGAGCTCAGTCCGCT CAGCGACAAGGAGAGTCAAGAGCCACCCAACGCAGAGAACCATCAGCCACGTCCTCCCTCCGACGAGCTCGACGA TGATCTTTACAACGTCGTCGTTATCGACTCCCCCAAGCGCCCTAAACGATCTTCTTTCATTCTCGAGGGTTTCGA GGACGACGACTTGTACAACGTTTGCGAGCCGAAGGAATTGTCCCCCATCCCAGGGCTTGGCTTATACGACGTGAA GAAGGACGCTACACCGACGCCGGTACCAGACTCCGACACCACCGCCAAGAAGGATCTTACGCCGACCCAGGAGCC AAAATCCGATACCACCCAGCAGGAGCCTACGCCAGCCCAGGAGCCAGAGTCCGACACCACTCAGAAGGATCCTGC GCCAACCCAAGAGGAAAAGTCCGATACCGCCAACCAGGATCCTACGCCGACTCAGGGGTCAGAGTCAGACACCAC CAAAAAGGATCTTACGCCGGCACAGAAGTCGAAATCCGACAACAAGAAGCAAGTATCCGAGGGCATCCCAGGATT GGTCTACTACCAAAAGCGACCACTCGCACGCGACGACGACGACACTGATTCCCCTGCTAAGAAGCTTAAAGCGAA TGACGACTCCGACCGCACTGTTATCCCCCAGAATAACCTCGGTTTCGGCTCCCGCATTCCCACCTTCAACTTCAA AGGTTCCTTCTTCCGCCAGCTCGAACTCAAAGAGCGCATCATGATCGTCAACCACCAACACCTTGGAGATCGACG AATGGACACCAGGCTTTTCATCTCGAGTGTTGTCCCCGCGACTCCCGGGGATCCCGACACTGCTGCCATTCGCAG ATCAGCACCGCGCGTCTTCGTGAAGCTACTCCATCCAAGCCCCGGATACGGAGTCAACACAACTGAAATTACCGC CTACAAGCGTATCGACAGGCTCAAGAACATACTCGACTCGGAGGTGAAGAAGTTCAGCATCTTCGTCTTATCCCT GGAAGCATACATATCCTGCTACGGTGGTCCAACTATGACCACCCAGATCTTCGTCATGCCACTTCTCAAGCGCGA TTTGTCTGCTGCGTTGCGAGAGCGAAACCAATACGACGGACCATCGGAAAACGTTCAACTGTGGATCGCGCAAAT TGCGGCCGGGGTCCATTTCCTCCACAATTGTGGTATCATGCATCGCGATCTCAAGTCAGAGAACATTCTGTTGGA TGCAGGCGACAACGCTTTGGTTACCGATTTCGGAAACTCTTTCGTTCAGGTCGACTGGATCAACGCCAAACTCGA TTATGACCTTTTGTACTCTCGTGCCAATTTAGGAACTCCTGGGTGGCTCGCTCCAGAGGTCGAACAAGGTTTGTG GTATGGCCCGGCAAGTGATTGGTGGGGTGTTGGCGCGATTATGCTCGAGGCCATGGTCCATCCGGCCAACTTCTA TATGGCGTTGTCTCAATTCCGGGCCACCCGACTGCTTTACCCGAGGAGCAGGAGCGGGGCAGACGCCAAGTTCGC TTTCGAGACTGAGCAAACCAGCGTCCCTTGCGCGTTTGCGAGAGAAGTCCTCTGGATGTTGCTTCACCACGACCC CCGCGACCGTTACCGTTATGACCAGCTTTACAGCCACACTTATTTCGCTGTTTACCGTGATGGCAGAGTGGTATC TATTTTCGACATTTTGGAGTTCGAAACCGCGGCACGACTGAACAAGGCCAAGCAAGAATCTGGACCAACGTCCGA AGATCCGACTGAGGATCATCATCAGCTTCCCTACAAGTGTGACCTTTGGAGGTTCGACCACCCTGGCACAAATGC GGAAGTGTGGCTCCCATCCTATTCGAAAATCCATCAAATCATCGACAAGGCTTATGAATAGTAGAGTAGTTAAGA CTCATACATACAAGCATCTTACATTTCTATTTCTCCGTTTCTTCGTGCATTTGTACTATATCTACATACCTCGAA AATTCGCACTCGCCACTAGGCGTGGAGTCGCAT |
| Length | 3183 |
Gene
| Sequence id | CopciAB_496994.T0 |
|---|---|
| Sequence |
>CopciAB_496994.T0 GCCGAAGTTTACTGCCAGCGTCGACGCAAGAATTCACCAAAACTCTGGCCATGGGGACGGGGACGTTTCAAAGTC TCATATCGGTGCTCTGGGATGTCTATCCTTTCAAGGTGCTCAACTGGATCTGTATGCTCGGAACTTGCAGTGCTG AACGACGCTCGACGACTTTCTGGACGACGACCCAACTGATTTTTCGGGCGGGTTTATCTGTTTCTCTTTTTCTGG GGGCAGGGACTGCACACAGAGCTCACACCACTGCACTGCTCAACGACCCAGGTGGATGGCGCTCTCGCTGTAGCC CATTGGCACCCAAATCTGTGCCATTCGCAATCGCGATCCCGCCATTGACGAGGACACGGTACGCCGAGAAGCCAC CCAAGCCATCGGAGAACGCCGAGTTTCGTGATCTTGCAACAAAGAGTGTGGGAACCCATTCCAAATGCGGTGGTG ATGTCGATGTCATGGACGCGCGCGTCTGTTGGACCATTGTCCGTTAGGGAGGCTCTCCTTCCAAATCCGTACTTT CGTCCGTCGCGACAACATGACAACTGGCCACTCCTTTGTGAGGGCGCACTCCATTGACATTATCTGCCTCGTGTT CTGGTTAAATGTACATACTTGGAAATTAGAATTCTGCGGACAATGGGCCCGGCCGGCGCTATAAAACAGACGACT CCTGCGATTTCACGGTTCTGCTTTTGCCCACTACCATCGCCTGTCCACCAGTCATATATCGCTCACCACATCCAA CCTTCGTGTCCCCTATACTCTCTTCTCATCTGCCACCATGGTACGAATCGCACTGTGGACCGTGATGAACAAACT TTCCCCTTCACGCGCGAAGGGCAAGTCCTCTCCTCGGTCGAGTCCAGTTCCATCCTCCTCCCCTTCGATCCACGT CGAAGAGCTCGGCGAGCTCAGCGACAAGGAGAATCAAGATCCATCTGTCGTCGAGAACTATCAACCATCTCCTCC CACCGTCGAGGTCATCGACTACACATCTCCACCCACCGTCGAGGTCGTCGACTACACATCTCCACCCACCGTCGA GGTCGTCGACTACACTCAAACATCACCTCCCACCGTCGAGGTCATCGATAATGCCACATCCGTCGAAGAACAGTC CTCTCCTCAGCCGAGTCCAGTTCCGTCCTCTTCGCCTTCCACCACCACCACTTCGGTCGAAGAGCTCAGTCCGCT CAGCGACAAGGAGAGTCAAGAGCCACCCAACGCAGAGAACCATCAGCCACGTCCTCCCTCCGACGAGCTCGACGA TGATCTTTACAACGTCGTCGTTATCGACTCCCCCAAGCGCCCTAAACGATCTTCTTTCATTCTCGAGGGTTTCGA GGACGACGACTTGTACAACGTTTGCGAGCCGAAGGAATTGTCCCCCATCCCAGGGCTTGGCTTATACGACGTGAA GAAGGACGCTACACCGACGCCGGTACCAGACTCCGACACCACCGCCAAGAAGGATCTTACGCCGACCCAGGAGCC AAAATCCGATACCACCCAGCAGGAGCCTACGCCAGCCCAGGAGCCAGAGTCCGACACCACTCAGAAGGATCCTGC GCCAACCCAAGAGGAAAAGTCCGATACCGCCAACCAGGATCCTACGCCGACTCAGGGGTCAGAGTCAGACACCAC CAAAAAGGATCTTACGCCGGCACAGAAGTCGAAATCCGACAACAAGAAGCAAGTATCCGAGGGCATCCCAGGATT GGTCTACTACCAAAAGCGACCACTCGCACGCGACGACGACGACACTGATTCCCCTGCTAAGAAGCTTAAAGCGAA TGACGACTCCGACCGCACTGTTATCCCCCAGAATAACCTCGGTTTCGGCTCCCGCATTCCCACCTTCAACTTCAA AGGTTCCTTCTTCCGCCAGCTCGAACTCAAAGAGCGCATCATGATCGTCAACCACCAACACCTTGGAGATCGACG AATGGACACCAGGCTTTTCATCTCGAGTGTTGTCCCCGCGACTCCCGGGGATCCCGACACTGCTGCCATTCGCAG ATCAGCACCGCGCGTCTTCGTGAAGCTACTCCATCCAAGCCCCGGATACGGAGTCAACACAACTGAAATTACCGC CTACAAGCGTATCGACAGGCTCAAGAACATACTCGACTCGGAGGTGAAGAAGTTCAGCATCTTCGTCTTATCCCT GGAAGCATACATATCCTGCTACGGTGGTCCAACTATGACCACCCAGATCTTCGTCATGGTATGTGATCTTTGGTA TTCTTCTATGTTCCCCTTTCTAACCACCTGTTGAACCACAGCCACTTCTCAAGCGCGATTTGTCTGCTGCGTTGC GAGAGCGAAACCAATACGACGGACCATCGGAAAACGTTCAACTGTGGATCGCGCAAATTGTAAGTCTGCCCTCCA CTACCACCGAGAACTTTCAGACTAACTCTGTCAAGGCGGCCGGGGTCCATTTCCTCCACAATTGTGGTATCATGC ATCGCGATCTCAAGTCAGAGAACATTCTGTTGGATGCAGGCGACAACGCTTTGGTTACCGATTTCGGAAACTCTT TCGTTCAGGTCGACTGGATCAACGCCAAACTCGATTATGACCTTTTGTACTCTCGTGCCAATTTAGGAACTCCTG GGTGGCTCGCTCCAGAGGTCGAACAAGGTTTGTGGTATGGCCCGGCAAGTGATTGGTGGGGTGTTGGCGCGATTA TGCTCGAGGCCATGGTCCATCCGGCCAACTTCTATGTACGTGACACGTTTACTCTGGCTTCTATCCGATGCTCAC CTGAATGATAGATGGCGTTGTCTCAATTCCGGGCCACCCGACTGCTTTACCCGAGGAGCAGGAGCGGGGCAGACG CCAAGTTCGCTTTCGAGACTGAGCAAACCAGCGTCCCTTGCGCGTTTGCGAGAGAAGTCCTCTGGATGGTATGTT TCCCATTCTGTGCGCTTTCCTGGTTTCGTGGTGCTGAATCGTGCTTTTCTCGTTTCAGTTGCTTCACCACGACCC CCGCGACCGTTACCGTTATGACCAGCTTTACAGCCACACTTATTTCGCTGTTTACCGTGATGGCAGAGTGGTATC TATTTTCGACATTTTGGAGTTCGAAAGTATGTGTTCCTTGGCCTTCTTCCTGTCATCGAGGCGGTTTGACCGACG CGTCCCCAGCCGCGGCACGACTGAACAAGGCCAAGCAAGAATCTGGACCAACGTCCGAAGATCCGACTGAGGATC ATCATCAGCTTCCCTACAAGTGTGACCTTTGGAGGTTCGACCACCCTGGCACAAATGCGGAAGTGTGGCTCCCAT CCTATTCGAAAATCCATCAAATCATCGACAAGGCTTATGAATAGTAGAGTAGTTAAGACTCATACATACAAGCAT CTTACATTTCTATTTCTCCGTTTCTTCGTGCATTTGTACTATATCTACATACCTCGAAAATTCGCACTCGCCACT AGGCGTGGAGTCGCAT |
| Length | 3466 |