CopciAB_497007
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_497007 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 497007 |
| Uniprot id | Functional description | protein kinase C activity | |
| Location | scaffold_9:1419076..1422075 | Strand | + |
| Gene length (nt) | 3000 | Transcript length (nt) | 2509 |
| CDS length (nt) | 2229 | Protein length (aa) | 742 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Pleurotus ostreatus PC9 | PleosPC9_1_46456 | 24.1 | 3.821E-26 | 115 |
| Pleurotus ostreatus PC15 | PleosPC15_2_153544 | 23.8 | 1.601E-25 | 113 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_110136 | 29.7 | 2.882E-24 | 109 |
| Grifola frondosa | Grifr_OBZ76670 | 33.9 | 5.825E-24 | 108 |
| Agrocybe aegerita | Agrae_CAA7268764 | 28.5 | 9.197E-22 | 101 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_497007.T0 |
| Description | protein kinase C activity |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 356 | 627 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008266 | Tyrosine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K18052 |
| K19662 |
EggNOG
| COG category | Description |
|---|---|
| T | protein kinase C activity |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
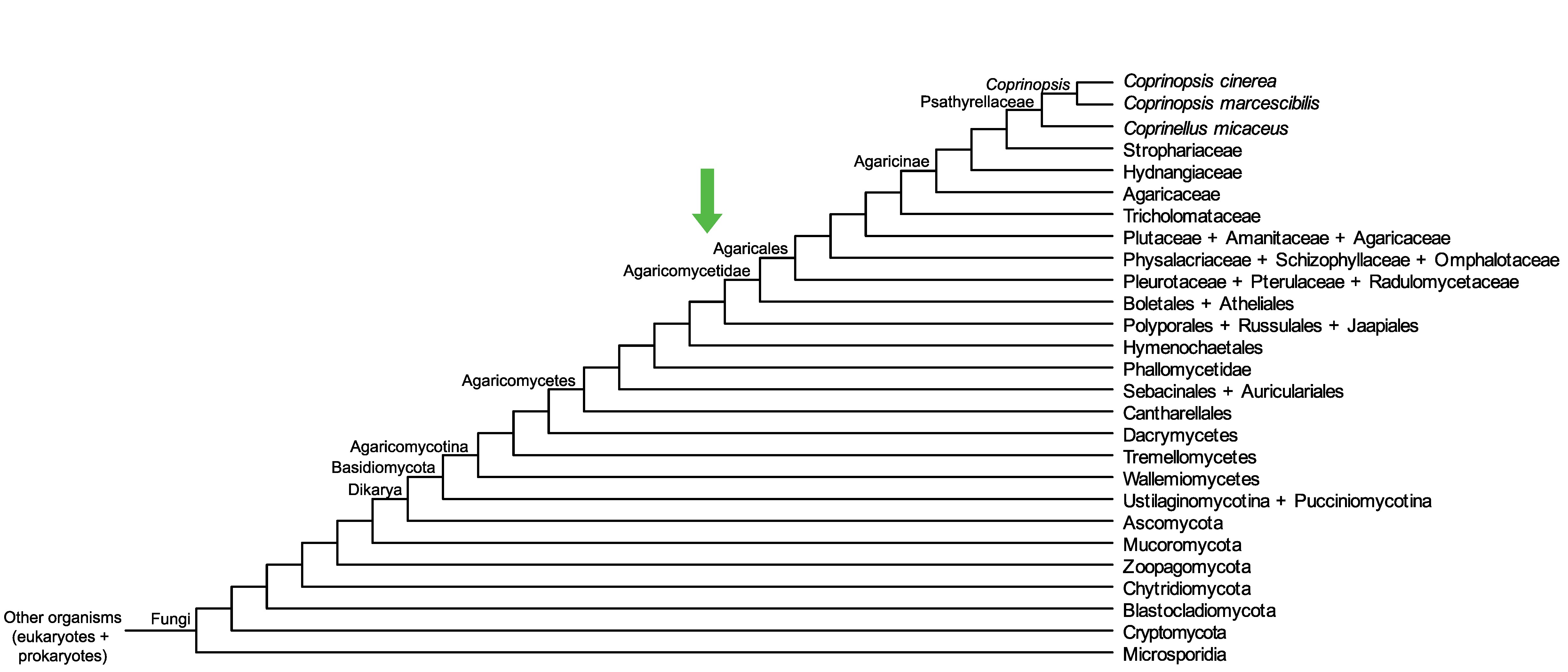
Conservation of CopciAB_497007 across fungi.
Arrow shows the origin of gene family containing CopciAB_497007.
Protein
| Sequence id | CopciAB_497007.T0 |
|---|---|
| Sequence |
>CopciAB_497007.T0 MEQRGWVNGLLESMWPRGGSIRFSEPFVTIYIAPRSTTRSRPRKPLATHIPPTCVLSSTQHAQHGQAGCPQSSSV IKGVFESAVFALSNAFEDRTNGAVPLSLDFVHIESYYGRALRNVPFVFAGVPWRGCAPFEWLGYKMGGRVGKAWY SPRYRGLDAMTPRRQISSSAEWEGSSPGYASSSGSSGYASSSSSSHASTSTAATSNSTSSIRKRIKRVFNRRDGK ATTEGPLFRYLVPPRPPVVNYHYTRVLPSGVPIFITPSHRKSTHPSYLPTYNSPLEFGLLESLNGRAEEEGRTMV TLAYFAKKDKREDPKLCDSTERMVVLKGNLFGFRSNRSRTRTGGEEETWEEELRERRKRAERKRVRTELRAMKRM AGGASRDGRGVFVVKADGCLMGPGVICIAMPYMKADLTEMYRNPKCRSYMARYGREWVMQLALGLSFIHDNGIIH RDICPQNILIDFNHTLKIADFDSAWVSQSSSSSPLALQSGVKYAHRLVGSKGYIAPEIISRCAPGKWTPAQREFL EGDHDVDERYAEFARYGKEVDWWSLGCVVAEMLTSAKDVGYAVLFGRDETLRLFVKSSPLERECVLRAYGLRGWA LSLVHGLLDPNPISRLGISGLRRHFYFMIRKDNPEEPIPNHPFEADRMDREGELMSLFDDPKKIVKNALVKEPLQ GQYRSWFEWESPHRESVTLGAYSPRGKPSAHVVQTVEEREEVDARTTEWWSWISPVGRWRGGLEGRD |
| Length | 742 |
Coding
| Sequence id | CopciAB_497007.T0 |
|---|---|
| Sequence |
>CopciAB_497007.T0 ATGGAACAGCGAGGTTGGGTCAATGGCCTCCTTGAAAGCATGTGGCCACGAGGCGGGTCTATCCGATTCTCCGAA CCCTTTGTGACGATATATATCGCGCCAAGGTCCACGACTAGAAGCAGACCAAGGAAGCCACTAGCAACGCACATC CCACCAACGTGTGTTCTCTCCTCGACCCAGCATGCCCAACATGGACAAGCAGGCTGTCCCCAGTCGTCTTCCGTC ATCAAAGGGGTTTTCGAGTCGGCCGTTTTTGCCCTTTCAAACGCGTTCGAAGATCGAACCAATGGGGCTGTTCCT TTGAGTCTGGATTTCGTGCATATCGAATCATATTATGGACGGGCTTTGAGGAACGTTCCCTTTGTTTTCGCAGGA GTGCCGTGGCGGGGATGTGCGCCCTTTGAATGGTTGGGGTATAAGATGGGTGGGAGGGTCGGTAAAGCCTGGTAT TCACCACGGTATAGAGGTCTCGACGCTATGACTCCTCGCAGGCAAATATCTTCCTCCGCAGAGTGGGAGGGCTCC AGCCCTGGTTATGCTTCGAGTTCTGGCTCTAGTGGCTATGCTTCATCGTCGAGTAGCAGCCACGCAAGTACCAGC ACCGCCGCCACTTCGAATTCAACCTCGAGCATCAGGAAGCGTATCAAAAGGGTCTTCAATAGAAGAGATGGCAAG GCGACAACGGAGGGTCCTCTATTCCGGTATCTCGTTCCTCCCAGACCACCCGTTGTCAATTATCACTACACTCGT GTCTTACCCTCTGGCGTCCCGATATTCATCACGCCCTCTCATCGCAAATCTACACACCCATCCTACCTCCCGACT TACAACTCGCCTCTCGAGTTCGGTCTACTAGAATCTCTAAATGGCAGGGCGGAGGAGGAAGGCAGGACGATGGTC ACCCTCGCCTACTTTGCAAAGAAGGACAAACGTGAAGATCCTAAACTGTGCGATTCAACGGAGAGGATGGTCGTG TTGAAGGGGAATTTGTTCGGATTCCGCTCAAATCGGTCTAGAACGAGAACTGGAGGGGAAGAGGAGACGTGGGAG GAAGAGCTACGGGAGAGGAGGAAGAGGGCCGAGAGGAAGAGGGTAAGAACGGAGCTGAGGGCTATGAAACGGATG GCGGGTGGTGCCAGTCGGGATGGGAGGGGTGTGTTTGTTGTAAAAGCGGACGGGTGCTTGATGGGCCCTGGTGTC ATTTGTATTGCCATGCCATATATGAAAGCAGATTTAACGGAGATGTACCGCAACCCCAAGTGTCGAAGCTACATG GCAAGGTATGGTCGGGAGTGGGTCATGCAGTTGGCGCTCGGCTTGAGCTTTATACACGACAACGGAATCATCCAC CGCGACATCTGTCCCCAAAACATCCTAATAGACTTCAACCACACGCTCAAAATCGCGGACTTCGATTCAGCTTGG GTCTCTCAAAGTTCCTCTTCCAGTCCTCTGGCACTCCAATCTGGAGTGAAGTACGCCCATCGGCTCGTTGGTTCG AAAGGGTACATCGCACCAGAGATTATCAGCCGTTGCGCACCCGGAAAGTGGACCCCTGCGCAACGCGAGTTCCTG GAAGGAGACCATGATGTAGACGAGCGGTATGCTGAGTTTGCGAGGTATGGGAAGGAGGTGGATTGGTGGTCGCTG GGATGTGTTGTCGCGGAGATGTTGACGAGTGCCAAGGACGTGGGGTATGCTGTGCTATTTGGCAGGGACGAAACG CTTAGGCTGTTCGTCAAGAGCAGTCCTCTGGAGAGGGAGTGTGTGTTGAGGGCGTATGGTCTACGTGGTTGGGCA TTGAGTCTTGTTCATGGTCTCCTTGATCCAAATCCAATATCTCGGCTCGGCATTTCGGGCCTTCGAAGGCATTTC TACTTCATGATTAGGAAGGATAACCCCGAGGAACCGATACCCAATCATCCATTCGAAGCCGACAGGATGGATAGG GAAGGGGAACTTATGAGCTTGTTCGACGATCCGAAAAAGATTGTAAAGAATGCTTTGGTCAAGGAGCCTCTACAA GGGCAGTATCGCTCGTGGTTCGAGTGGGAGTCACCACATAGGGAGAGCGTGACTTTGGGTGCGTACTCGCCTCGA GGCAAGCCTTCTGCGCATGTTGTGCAGACAGTGGAAGAGAGGGAAGAGGTGGATGCGAGGACGACAGAGTGGTGG AGTTGGATCAGTCCTGTTGGAAGGTGGAGGGGCGGGCTTGAGGGGCGGGATTAG |
| Length | 2229 |
Transcript
| Sequence id | CopciAB_497007.T0 |
|---|---|
| Sequence |
>CopciAB_497007.T0 AGCCACTACCTTGTCGGCTGGCGCCAACGTCGAACACATTGGATAGCCCCTGATAAATGGTTATCGACCGTTTCA GCGTTGAAGCTTTGTTATATCTGGATGGGCGGTGACAATGGCCCTGGATGGAACAGCGAGGTTGGGTCAATGGCC TCCTTGAAAGCATGTGGCCACGAGGCGGGTCTATCCGATTCTCCGAACCCTTTGTGACGATATATATCGCGCCAA GGTCCACGACTAGAAGCAGACCAAGGAAGCCACTAGCAACGCACATCCCACCAACGTGTGTTCTCTCCTCGACCC AGCATGCCCAACATGGACAAGCAGGCTGTCCCCAGTCGTCTTCCGTCATCAAAGGGGTTTTCGAGTCGGCCGTTT TTGCCCTTTCAAACGCGTTCGAAGATCGAACCAATGGGGCTGTTCCTTTGAGTCTGGATTTCGTGCATATCGAAT CATATTATGGACGGGCTTTGAGGAACGTTCCCTTTGTTTTCGCAGGAGTGCCGTGGCGGGGATGTGCGCCCTTTG AATGGTTGGGGTATAAGATGGGTGGGAGGGTCGGTAAAGCCTGGTATTCACCACGGTATAGAGGTCTCGACGCTA TGACTCCTCGCAGGCAAATATCTTCCTCCGCAGAGTGGGAGGGCTCCAGCCCTGGTTATGCTTCGAGTTCTGGCT CTAGTGGCTATGCTTCATCGTCGAGTAGCAGCCACGCAAGTACCAGCACCGCCGCCACTTCGAATTCAACCTCGA GCATCAGGAAGCGTATCAAAAGGGTCTTCAATAGAAGAGATGGCAAGGCGACAACGGAGGGTCCTCTATTCCGGT ATCTCGTTCCTCCCAGACCACCCGTTGTCAATTATCACTACACTCGTGTCTTACCCTCTGGCGTCCCGATATTCA TCACGCCCTCTCATCGCAAATCTACACACCCATCCTACCTCCCGACTTACAACTCGCCTCTCGAGTTCGGTCTAC TAGAATCTCTAAATGGCAGGGCGGAGGAGGAAGGCAGGACGATGGTCACCCTCGCCTACTTTGCAAAGAAGGACA AACGTGAAGATCCTAAACTGTGCGATTCAACGGAGAGGATGGTCGTGTTGAAGGGGAATTTGTTCGGATTCCGCT CAAATCGGTCTAGAACGAGAACTGGAGGGGAAGAGGAGACGTGGGAGGAAGAGCTACGGGAGAGGAGGAAGAGGG CCGAGAGGAAGAGGGTAAGAACGGAGCTGAGGGCTATGAAACGGATGGCGGGTGGTGCCAGTCGGGATGGGAGGG GTGTGTTTGTTGTAAAAGCGGACGGGTGCTTGATGGGCCCTGGTGTCATTTGTATTGCCATGCCATATATGAAAG CAGATTTAACGGAGATGTACCGCAACCCCAAGTGTCGAAGCTACATGGCAAGGTATGGTCGGGAGTGGGTCATGC AGTTGGCGCTCGGCTTGAGCTTTATACACGACAACGGAATCATCCACCGCGACATCTGTCCCCAAAACATCCTAA TAGACTTCAACCACACGCTCAAAATCGCGGACTTCGATTCAGCTTGGGTCTCTCAAAGTTCCTCTTCCAGTCCTC TGGCACTCCAATCTGGAGTGAAGTACGCCCATCGGCTCGTTGGTTCGAAAGGGTACATCGCACCAGAGATTATCA GCCGTTGCGCACCCGGAAAGTGGACCCCTGCGCAACGCGAGTTCCTGGAAGGAGACCATGATGTAGACGAGCGGT ATGCTGAGTTTGCGAGGTATGGGAAGGAGGTGGATTGGTGGTCGCTGGGATGTGTTGTCGCGGAGATGTTGACGA GTGCCAAGGACGTGGGGTATGCTGTGCTATTTGGCAGGGACGAAACGCTTAGGCTGTTCGTCAAGAGCAGTCCTC TGGAGAGGGAGTGTGTGTTGAGGGCGTATGGTCTACGTGGTTGGGCATTGAGTCTTGTTCATGGTCTCCTTGATC CAAATCCAATATCTCGGCTCGGCATTTCGGGCCTTCGAAGGCATTTCTACTTCATGATTAGGAAGGATAACCCCG AGGAACCGATACCCAATCATCCATTCGAAGCCGACAGGATGGATAGGGAAGGGGAACTTATGAGCTTGTTCGACG ATCCGAAAAAGATTGTAAAGAATGCTTTGGTCAAGGAGCCTCTACAAGGGCAGTATCGCTCGTGGTTCGAGTGGG AGTCACCACATAGGGAGAGCGTGACTTTGGGTGCGTACTCGCCTCGAGGCAAGCCTTCTGCGCATGTTGTGCAGA CAGTGGAAGAGAGGGAAGAGGTGGATGCGAGGACGACAGAGTGGTGGAGTTGGATCAGTCCTGTTGGAAGGTGGA GGGGCGGGCTTGAGGGGCGGGATTAGACAAGCGCACCATAGCTGAAGAGGGGCTCGGTAGCTTATTGGGCTATTG CGATTCCGTTCCGTGCAGCACAATAGTACTTTAGAGCTTCTGGCATATTACCTTTCGACAAATTACTTTAAATTC TGCTACTTGCGTAGAATTGACATTGGTTTTATCC |
| Length | 2509 |
Gene
| Sequence id | CopciAB_497007.T0 |
|---|---|
| Sequence |
>CopciAB_497007.T0 AGCCACTACCTTGTCGGCTGGCGCCAACGTCGAACACATTGGATAGCCCCTGATAAATGGTTATCGACCGTTTCA GCGTTGAAGCTTTGTTATATCTGGATGGGCGGTGACAATGGCCCTGGATGGAACAGCGAGGTTGGGTCAATGGCC TCCTTGAAAGCATGTGGCCACGAGGCGGGTCTATCCGATTCTCCGAACCCTTTGTGACGATATATATCGCGCCAA GGTCCACGGTACGTCTTCCCCGTCTTCCTCCTAGTACGCACCCACAGATCTTGATACGTATACCTTTGTAGACTA GAAGCAGACCAAGGAAGCCACTAGCAACGCACATCCCACCAACGTGTGTTCTCTCCTCGACCCAGCATGCCCAAC ATGGACAAGCAGGCTGTCCCCAGTCGTCTTCCGTCATCAAAGGGGTTTTCGAGTCGGCCGTTTTTGCCCTTTCAA ACGCGTTCGAAGATCGAACCAATGGGGCTGTTCCTTTGAGTCTGGATTTCGTGCATATCGAATCATATTATGGAC GGGCTTTGAGGAACGTTCCCTTTGTTTTCGCAGGAGTGCCGTGGCGGGGATGTGCGCCCTTTGAATGGTTGGGGT ATAAGATGGGTGGGAGGGTCGGTAAAGCCTGGTATTCACCACGGTATAGAGGTCTCGACGCTATGACTCCTCGCA GGCAAATATCTTCCTCCGCAGAGTGGGAGGGCTCCAGCCCTGGTTATGCTTCGAGTTCTGGCTCTAGTGGCTATG GTCAGTTCAACCCCATCATTTTGTCCTGCCACATGAATTGACGGTATCCTTTTCCGTGGATAGCTTCATCGTCGA GTAGCAGCCACGCAAGTACCAGCACCGCCGCCACTTCGAATTCAACCTCGAGCATCAGGAAGCGTATCAAAAGGG TCTTCAATAGAAGAGATGGCAAGGCGACAACGGAGGGTCCTCTATTCCGGTATCTCGTTCGTAAGTACCTAACGT GGTACCTCGAACTCACTAAAGACGAACCAACGATTTCAACAGCTCCCAGACCACCCGTTGTCAATTATCACTACA CTCGTGTCTTACCCTCTGGCGTCCCGATATTCATCACGCCCTCTCATCGCAAATCTACACACCCATCCTACCTCC CGACTTACAACTCGCCTCTCGAGTTCGGTCTACTAGAATCTCTAAATGGCAGGGCGGAGGAGGAAGGCAGGACGA TGGTCACCCTCGCCTACTTTGCAAAGAAGGACAAACGTGAAGATCCTAAACTGTGCGATTCAACGGAGAGGATGG TCGTGTTGAAGGGGAATTTGTTCGGATTCCGCTCAAATCGGTCTAGAACGAGAACTGGAGGGGAAGAGGAGACGT GGGAGGAAGAGCTACGGGAGAGGAGGAAGAGGGCCGAGAGGAAGAGGGTAAGAACGGAGCTGAGGGCTATGAAAC GGATGGCGGGTGGTGCCAGTCGGGATGGGAGGGGTGTGTTTGTTGTAAAAGCGGACGGGTGCTTGATGGGCCCTG GTGTCATTTGTATTGCCATGGTATGCCTATTCGAAGAGTGAGAGGGCTGATTGACTGACTTCAAGTTATTGACTT CCAGCCATATATGAAAGCAGATTTAACGGAGATGTACCGCAACCCCAAGTGTCGAAGCTACATGGCAAGGTATGG TCGGGAGTGGGTCATGCAGTTGGTGAGTCTCCACCTTGACGAGGTGGTGTAACAGTAACTAACTGGCTGATGTAC CCCTTCCTTTCTACTGAACAGGCGCTCGGCTTGAGCTTTATACACGACAACGGAATCATCCACCGCGACATCTGT CCCCAAAACATCCTAATAGACTTCAACCACACGCTCAAAATCGCGGACTTCGATTCAGCTTGGGTCTCTCAAAGT TCCTCTTCCAGTCCTCTGGCACTCCAATCTGGAGTGAAGTACGCCCATCGGCTCGTTGGTTCGAAAGGGTACATC GCACCAGAGATTATCAGCCGTTGCGCACCCGGAAAGTGGACCCCTGCGCAACGCGAGTTCCTGGAAGGAGACCAT GATGTAGACGAGCGGTATGCTGAGTTTGCGAGGTATGGGAAGGAGGTGGATTGGTGGTCGCTGGGATGTGTTGTC GCGGAGATGTTGACGAGTGCCAAGGACGTGGGGTATGCTGTCAGTGTTCTTTGAATTATCAATTGGCCGTGGATG AGACGAGGGCTAATCACGAAGCTAATGTAGGTGCTATTTGGCAGGGACGAAACGCTTAGGCTGTTCGTCAAGAGC AGTCCTCTGGAGAGGGAGTGTGTGTTGAGGGCGTATGGTCTACGTGGTTGGGCATTGAGTCTTGTTCATGGTGTA CGTCTCCCTCGAATACGCAGCCAGGGGGATGGCACGCTGACTTTGTCTCCAGCTCCTTGATCCAAATCCAATATC TCGGCTCGGCATTTCGGGCCTTCGAAGGCATTTCTACTTCATGATTAGGAAGGATAACCCCGAGGAACCGATACC CAATCATCCATTCGAAGCCGACAGGATGGATAGGGAAGGGGAACTTATGAGCTTGTTCGACGATCCGAAAAAGAT TGTAAAGAATGGTACATTATTCTGACACGGTATGTGACGAGACGAAACTAATTTCCTGGCTTCAGCTTTGGTCAA GGAGCCTCTACAAGGGCAGTATCGCTCGTGGTTCGAGTGGGAGTCACCACATAGGGAGAGCGTGACTTTGGGTGC GTACTCGCCTCGAGGCAAGCCTTCTGCGCATGTTGTGCAGACAGTGGAAGAGAGGGAAGAGGTGGATGCGAGGAC GACAGAGTGGTGGAGTTGGATCAGTCCTGTTGGAAGGTGGAGGGGCGGGCTTGAGGGGCGGGATTAGACAAGCGC ACCATAGCTGAAGAGGGGCTCGGTAGCTTATTGGGCTATTGCGATTCCGTTCCGTGCAGCACAATAGTACTTTAG AGCTTCTGGCATATTACCTTTCGACAAATTACTTTAAATTCTGCTACTTGCGTAGAATTGACATTGGTTTTATCC |
| Length | 3000 |