CopciAB_500402
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_500402 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 500402 |
| Uniprot id | Functional description | alpha/beta hydrolase fold | |
| Location | scaffold_7:638484..639868 | Strand | - |
| Gene length (nt) | 1385 | Transcript length (nt) | 1270 |
| CDS length (nt) | 1164 | Protein length (aa) | 387 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262899 | 48.9 | 3.38E-106 | 347 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_84880 | 46.5 | 4.766E-100 | 329 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_207012 | 46.9 | 4.153E-99 | 326 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1045533 | 45.4 | 5.196E-98 | 323 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1450099 | 44.9 | 2.986E-97 | 321 |
| Pleurotus ostreatus PC9 | PleosPC9_1_58363 | 45.2 | 1.214E-96 | 319 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_74194 | 46.7 | 2.467E-96 | 318 |
| Schizophyllum commune H4-8 | Schco3_1124648 | 45.3 | 4.562E-93 | 309 |
| Lentinula edodes NBRC 111202 | Lenedo1_465944 | 42 | 4.672E-91 | 303 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_14423 | 42.3 | 1.024E-90 | 302 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14647 | 46 | 7.228E-76 | 259 |
| Grifola frondosa | Grifr_OBZ71192 | 50.1 | 1.396E-60 | 214 |
| Lentinula edodes B17 | Lened_B_1_1_9020 | 37.9 | 1.27E-59 | 211 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_500402.T0 |
| Description | alpha/beta hydrolase fold |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07859 | alpha/beta hydrolase fold | IPR013094 | 44 | 170 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR029058 | Alpha/Beta hydrolase fold |
| IPR013094 | Alpha/beta hydrolase fold-3 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016787 | hydrolase activity | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | alpha/beta hydrolase fold |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| CE | CE10 |
Transcription factor
| Group |
|---|
| No records |
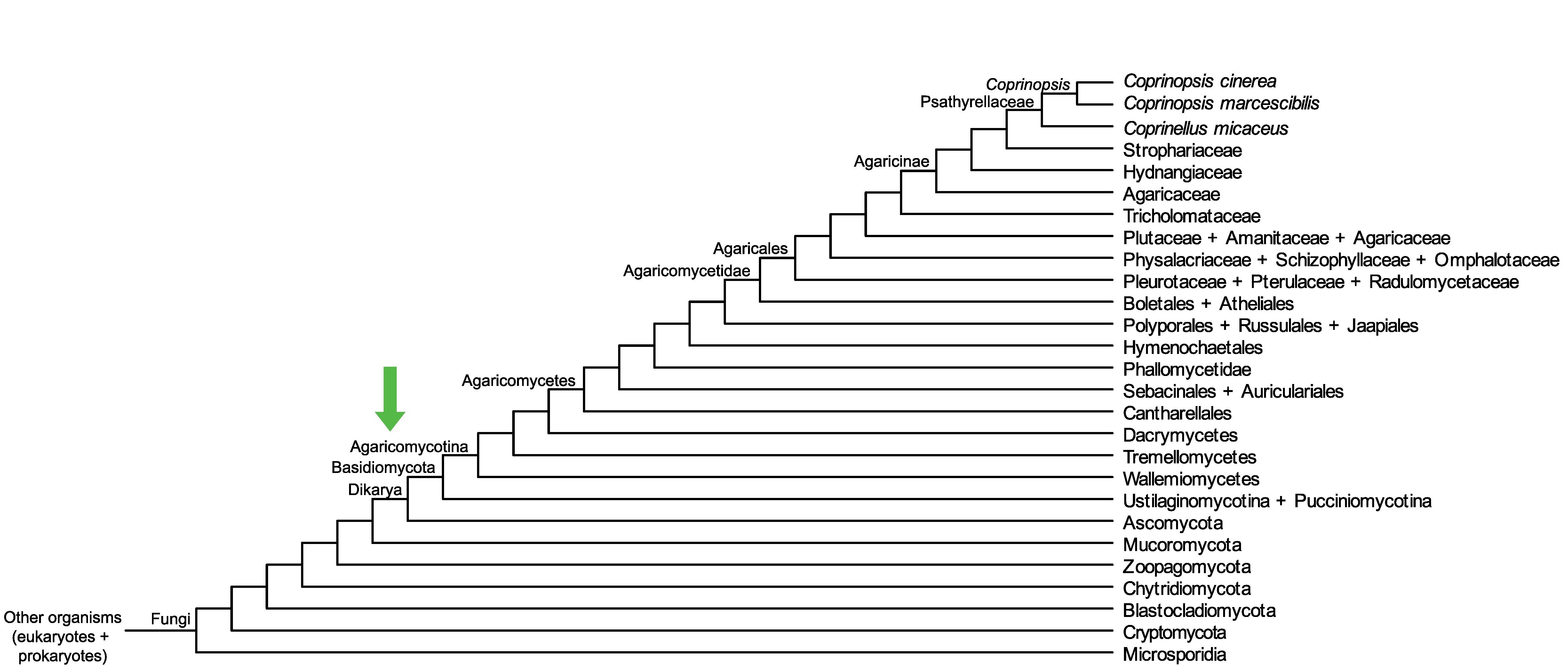
Conservation of CopciAB_500402 across fungi.
Arrow shows the origin of gene family containing CopciAB_500402.
Protein
| Sequence id | CopciAB_500402.T0 |
|---|---|
| Sequence |
>CopciAB_500402.T0 MSAMSSEPLTVTYKRRGDLNILSDVWQPESGPTPNGDKITVPAVVFFHGGGLTVGNRRSWFPEWLCKRFTKLGYL FISADYSLIPPATGHDIVEDIKDLWSFILDPHNIFSFVNGGTQKDFVVQPDKIVVAGTSAGGLCAYLCAMHCVDP PPVALLSIYGMGGDFFTPHYLHEKHAPFFRGRELLDPSEFQEFVYPFQEGDSTFVSDSPLSHHPPTHPIPGYPSN PRMLLARLYLQLGQFLDYYTGQHSPSLSRQLREIYDSYHQEHTQPSTLKGASPPLDGGDDDSSRLWEAIESCLGQ HRNLFPQVNVVKGEGRWPRTVFLHGTADTAVPIGESRSMESQLKAIGVPTQLFEVEEEEHSFDLVPNAEERFGVI FDKVTAFLHGVE |
| Length | 387 |
Coding
| Sequence id | CopciAB_500402.T0 |
|---|---|
| Sequence |
>CopciAB_500402.T0 ATGTCAGCGATGTCTTCTGAGCCCCTCACGGTCACGTACAAAAGAAGAGGAGACTTGAACATTCTTTCGGACGTA TGGCAACCTGAAAGCGGACCGACGCCGAACGGGGACAAGATTACAGTTCCGGCAGTGGTGTTCTTCCATGGCGGT GGGCTGACAGTCGGGAATAGACGTAGCTGGTTTCCTGAGTGGTTATGTAAACGTTTCACCAAGCTTGGATATCTT TTTATTTCAGCGGACTACAGTCTGATCCCTCCGGCAACTGGACATGACATCGTGGAGGACATCAAGGACTTGTGG TCCTTCATACTCGATCCTCACAATATCTTCTCCTTCGTAAATGGGGGGACGCAGAAGGATTTTGTCGTGCAGCCG GACAAAATCGTCGTAGCGGGAACAAGCGCTGGTGGTCTTTGTGCCTATCTGTGCGCGATGCACTGTGTTGATCCT CCTCCCGTGGCGCTGCTCTCGATCTACGGAATGGGTGGAGATTTCTTCACGCCTCACTATCTACATGAGAAACAT GCACCGTTCTTCCGAGGAAGGGAGCTTCTCGATCCCTCCGAGTTCCAGGAGTTTGTGTATCCGTTCCAAGAGGGG GACTCAACATTCGTATCTGACTCACCCCTCTCACACCACCCTCCAACCCACCCCATCCCCGGCTACCCTTCCAAT CCCCGGATGCTTCTCGCACGGCTGTACCTGCAGCTCGGTCAATTTCTGGATTACTACACAGGCCAGCATTCACCC AGTCTAAGCCGCCAGCTCCGAGAGATTTATGACTCTTATCATCAGGAACATACCCAGCCTTCGACATTGAAGGGT GCGTCACCTCCCCTTGACGGGGGAGATGACGATTCCTCTCGGCTTTGGGAGGCAATCGAGTCATGCCTTGGTCAA CATCGAAACCTGTTCCCGCAGGTTAACGTTGTGAAAGGGGAAGGTCGGTGGCCGAGGACGGTCTTCCTTCATGGA ACAGCAGATACGGCTGTCCCGATAGGCGAATCGAGGAGCATGGAAAGTCAACTGAAGGCAATTGGAGTGCCTACC CAGCTTTTCGAAGTTGAAGAAGAGGAGCATTCCTTTGATCTAGTACCTAACGCAGAGGAAAGGTTTGGCGTTATT TTTGATAAAGTGACGGCCTTCCTACATGGTGTAGAGTAA |
| Length | 1164 |
Transcript
| Sequence id | CopciAB_500402.T0 |
|---|---|
| Sequence |
>CopciAB_500402.T0 ATCTTCCTTCTCTTCATTCACATCCATGTCAGCGATGTCTTCTGAGCCCCTCACGGTCACGTACAAAAGAAGAGG AGACTTGAACATTCTTTCGGACGTATGGCAACCTGAAAGCGGACCGACGCCGAACGGGGACAAGATTACAGTTCC GGCAGTGGTGTTCTTCCATGGCGGTGGGCTGACAGTCGGGAATAGACGTAGCTGGTTTCCTGAGTGGTTATGTAA ACGTTTCACCAAGCTTGGATATCTTTTTATTTCAGCGGACTACAGTCTGATCCCTCCGGCAACTGGACATGACAT CGTGGAGGACATCAAGGACTTGTGGTCCTTCATACTCGATCCTCACAATATCTTCTCCTTCGTAAATGGGGGGAC GCAGAAGGATTTTGTCGTGCAGCCGGACAAAATCGTCGTAGCGGGAACAAGCGCTGGTGGTCTTTGTGCCTATCT GTGCGCGATGCACTGTGTTGATCCTCCTCCCGTGGCGCTGCTCTCGATCTACGGAATGGGTGGAGATTTCTTCAC GCCTCACTATCTACATGAGAAACATGCACCGTTCTTCCGAGGAAGGGAGCTTCTCGATCCCTCCGAGTTCCAGGA GTTTGTGTATCCGTTCCAAGAGGGGGACTCAACATTCGTATCTGACTCACCCCTCTCACACCACCCTCCAACCCA CCCCATCCCCGGCTACCCTTCCAATCCCCGGATGCTTCTCGCACGGCTGTACCTGCAGCTCGGTCAATTTCTGGA TTACTACACAGGCCAGCATTCACCCAGTCTAAGCCGCCAGCTCCGAGAGATTTATGACTCTTATCATCAGGAACA TACCCAGCCTTCGACATTGAAGGGTGCGTCACCTCCCCTTGACGGGGGAGATGACGATTCCTCTCGGCTTTGGGA GGCAATCGAGTCATGCCTTGGTCAACATCGAAACCTGTTCCCGCAGGTTAACGTTGTGAAAGGGGAAGGTCGGTG GCCGAGGACGGTCTTCCTTCATGGAACAGCAGATACGGCTGTCCCGATAGGCGAATCGAGGAGCATGGAAAGTCA ACTGAAGGCAATTGGAGTGCCTACCCAGCTTTTCGAAGTTGAAGAAGAGGAGCATTCCTTTGATCTAGTACCTAA CGCAGAGGAAAGGTTTGGCGTTATTTTTGATAAAGTGACGGCCTTCCTACATGGTGTAGAGTAATCTGGCTCCTC CAGGACTCCAGCGGTTTCCTGAAACTTTCAATGTTTTTCCGAATCTAAGAATAGAGATGATTCGCCGAGA |
| Length | 1270 |
Gene
| Sequence id | CopciAB_500402.T0 |
|---|---|
| Sequence |
>CopciAB_500402.T0 ATCTTCCTTCTCTTCATTCACATCCATGTCAGCGATGTCTTCTGAGCCCCTCACGGTCACGTACAAAAGAAGAGG AGACTTGAACATTCTTTCGGACGTATGGCAACCTGAAAGCGGACCGACGCCGAACGGGGACAAGATTACAGTTCC GGCAGTGGTGTTCTTCCATGGCGGTGGGCTGACAGTCGGGAATAGACGTAGCTGGTTTCCTGAGTGGTTATGTAG TATGTCTTTAAAATCGCATCAGGGAACGGTTTGAATCCTCACATCTCAAGAAAGAACGTTTCACCAAGCTTGGAT ATCTTTTTATTTCAGCGGACTACAGTCTGATCCCTCCGGCAACTGGACATGACATCGTGGAGGACATCAAGGACT TGTGGTCCTTCATACTCGATCCTCACAATATCTTCTCCTTCGTAAATGGGGGGACGCAGAAGGATTTTGTCGTGC AGCCGGACAAAATCGTCGTAGCGGGAACAAGCGCTGGTGGTCTTTGTGCCTATCTGTGCGCGATGCACTGTGTTG ATCCTCCTCCCGTGGCGCTGCTCTCGATCTACGGAATGGGTGGAGATTTCTTCGTGAGTTATACTTTCGCTGTCC GTTACTGCATGGTGATTGATGATACGCTGGCGAGACAGACGCCTCACTATCTACATGAGAAACATGCACCGTTCT TCCGAGGAAGGGAGCTTCTCGATCCCTCCGAGTTCCAGGAGTTTGTGTATCCGTTCCAAGAGGGGGACTCAACAT TCGTATCTGACTCACCCCTCTCACACCACCCTCCAACCCACCCCATCCCCGGCTACCCTTCCAATCCCCGGATGC TTCTCGCACGGCTGTACCTGCAGCTCGGTCAATTTCTGGATTACTACACAGGCCAGCATTCACCCAGTCTAAGCC GCCAGCTCCGAGAGATTTATGACTCTTATCATCAGGAACATACCCAGCCTTCGACATTGAAGGGTGCGTCACCTC CCCTTGACGGGGGAGATGACGATTCCTCTCGGCTTTGGGAGGCAATCGAGTCATGCCTTGGTCAACATCGAAACC TGTTCCCGCAGGTTAACGTTGTGAAAGGGGAAGGTCGGTGGCCGAGGACGGTCTTCCTTCATGGAACAGCAGATA CGGCTGTCCCGATAGGCGAATCGAGGAGCATGGAAAGTCAACTGAAGGCAATTGGAGTGCCTACCCAGCTTTTCG AAGTTGAAGAAGAGGAGCATTCCTTTGATCTAGTACCTAACGCAGAGGAAAGGTTTGGCGTTATTTTTGATAAAG TGACGGCCTTCCTACATGGTGTAGAGTAATCTGGCTCCTCCAGGACTCCAGCGGTTTCCTGAAACTTTCAATGTT TTTCCGAATCTAAGAATAGAGATGATTCGCCGAGA |
| Length | 1385 |