CopciAB_500464
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_500464 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | exgD | Synonyms | 500464 |
| Uniprot id | Functional description | Glycoside hydrolase family 5 protein | |
| Location | scaffold_7:322465..325868 | Strand | + |
| Gene length (nt) | 3404 | Transcript length (nt) | 3035 |
| CDS length (nt) | 2301 | Protein length (aa) | 766 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | NCU03914 |
| Schizosaccharomyces pombe | exg2 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| 0 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | exgD |
|---|---|
| Protein id | CopciAB_500464.T0 |
| Description | Glycoside hydrolase family 5 protein |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 288 | 490 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 111 | 133 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Glycoside hydrolase family 5 protein |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_9 |
Transcription factor
| Group |
|---|
| No records |
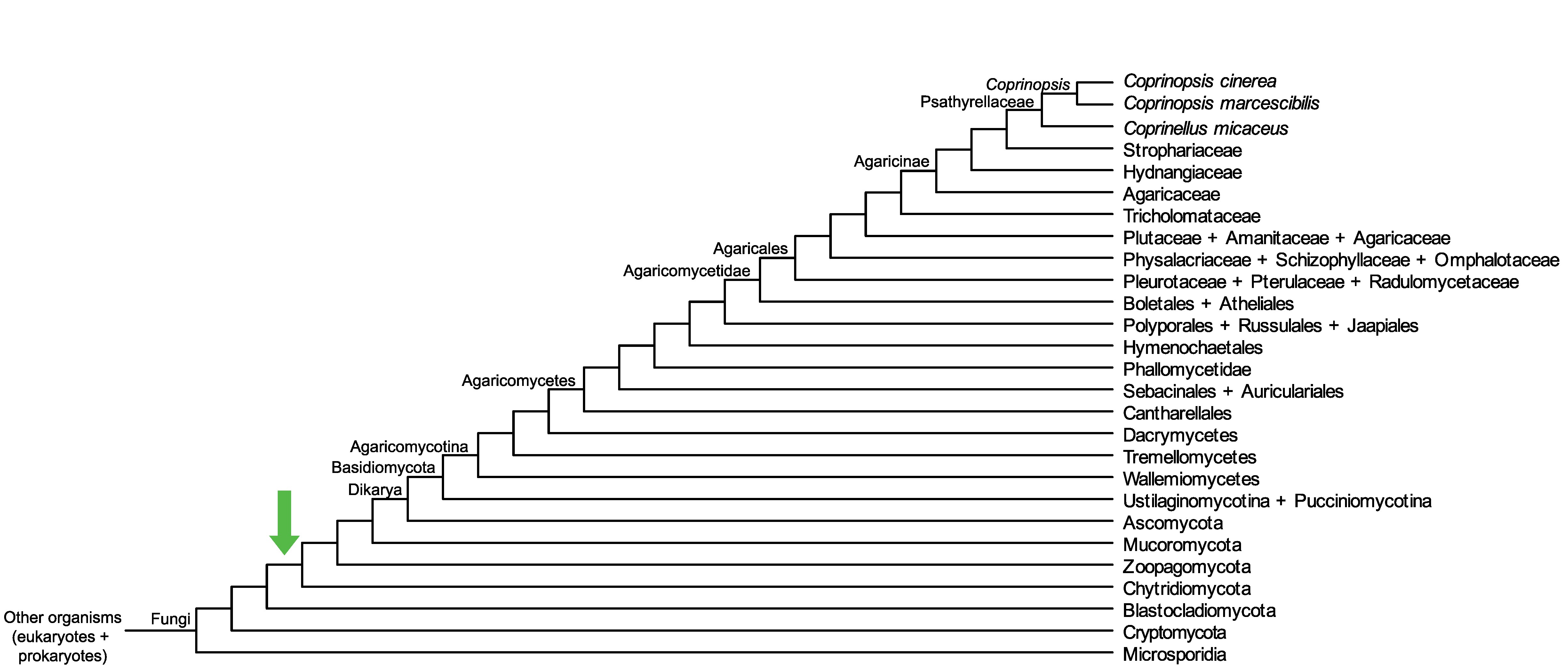
Conservation of CopciAB_500464 across fungi.
Arrow shows the origin of gene family containing CopciAB_500464.
Protein
| Sequence id | CopciAB_500464.T0 |
|---|---|
| Sequence |
>CopciAB_500464.T0 MSHNPQQSVTGGGAYDPLPLTGGEGHQPRFMGQYDAAASSSQQSLQAPSEYGSAHRLNDANDVNAAPYRDDPNAV PMSPLGASAAAMNEKSGGGGGSGGEGGRSRRKMVIIGAIVALIIIILAIILPVYFTVIKKDDNDSAGGGDRDSSN ADEGKDGSGTARPSGSGSPEPDVELPPVTGGDGSIITMEDGTTFTYRNPFGGYWYYDPNDPFNNGARAQSWTPAL NETFNYGVDRIRGVNVGGWLVTEPFISPALYERYLNDPAGPAIDEYDLSLRMRADTANGGINQLEEHYRTFITEK DFADIAGAGLNYVRIPIGWWAVETRGDEPHLEGVSWNYFLKAIKWARKYGLRINLDLHAVPGSQNAWNHSGKFGS IGFLHGPMGYANAQRTLDIIRVLAEFISQPQYRDVVTMFGILNEPLGDPMGFDALARFYMEAYTIIRRAGGIGEG NGPWVSLHDGFFGRDRWSGVFPNADRLALDVHPYLGFGSQSSAPMSSYADTPCRAWGRLVNNSMAAFGLTTAGEW SNAVNDCGLYLNGVNLGTRYEGTYTPGSWPRVGDCRDWTDWTRYTPELKREIRQFTLASMDALQHFFFWTWRIGD SLASGRVETPAWSYVLGLQEGWMPTDPRDADGVCNNIAPWTPPLSVGSGAIPASVTSALSWPPPAISNGGPIEVL PSYTQTGAIPTLTATPVEAAPGETITRTVDVGSGWNNPEDSEGYVAEVEGCSYLDPWVTPAAAPPSPLCVGVARR GMLGRGVVEARATPAP |
| Length | 766 |
Coding
| Sequence id | CopciAB_500464.T0 |
|---|---|
| Sequence |
>CopciAB_500464.T0 ATGTCGCATAACCCCCAGCAATCTGTAACGGGCGGGGGCGCCTACGACCCGCTCCCCCTCACCGGCGGCGAAGGA CACCAACCGCGGTTTATGGGCCAATACGACGCTGCCGCCTCGTCGTCGCAGCAGAGCCTGCAAGCGCCATCCGAG TATGGCTCGGCCCACCGCCTCAACGACGCCAACGATGTCAATGCAGCACCTTACCGCGACGACCCGAATGCCGTG CCCATGTCGCCGTTGGGTGCGTCTGCTGCCGCTATGAACGAGAAGTCCGGAGGAGGAGGGGGGAGTGGCGGCGAG GGTGGACGGTCGCGCCGCAAGATGGTGATCATCGGCGCCATCGTCGCGCTCATCATCATCATCCTAGCCATTATC CTCCCCGTCTACTTCACCGTGATCAAGAAGGACGATAATGATAGTGCTGGTGGTGGCGACCGGGATAGCAGTAAC GCGGACGAGGGTAAGGATGGGAGTGGGACGGCTAGGCCGAGTGGGAGTGGGAGTCCTGAGCCGGACGTTGAGCTG CCCCCTGTTACTGGCGGCGATGGGAGTATCATTACGATGGAGGATGGTACGACTTTTACGTATCGCAATCCGTTT GGAGGGTATTGGTATTATGATCCCAATGATCCTTTTAATAATGGGGCGAGGGCGCAGTCGTGGACTCCGGCGTTG AATGAGACGTTTAACTATGGTGTTGATAGGATTCGGGGTGTTAATGTTGGTGGTTGGCTTGTTACTGAGCCATTC ATTTCACCGGCTTTGTATGAGCGATATCTCAACGACCCTGCTGGACCTGCCATCGATGAATACGACCTCAGCTTG CGCATGCGCGCTGATACAGCCAACGGAGGCATCAACCAACTCGAGGAACATTATAGGACTTTCATCACTGAGAAG GACTTTGCTGATATCGCTGGTGCCGGTCTGAACTATGTCCGTATTCCTATTGGTTGGTGGGCCGTCGAGACGCGT GGCGATGAGCCTCACTTGGAGGGTGTTTCTTGGAACTACTTCCTCAAGGCGATCAAGTGGGCTAGGAAATACGGT CTCAGGATCAACCTCGATCTTCACGCTGTACCCGGAAGCCAGAACGCCTGGAACCACTCGGGCAAGTTTGGCAGC ATCGGATTCTTGCACGGTCCTATGGGTTACGCCAATGCGCAGAGGACGCTCGATATTATCCGTGTCCTTGCCGAG TTCATCTCGCAGCCTCAGTATCGGGACGTGGTCACCATGTTTGGTATCCTCAATGAGCCTTTGGGTGATCCTATG GGCTTTGATGCTCTAGCTCGATTCTACATGGAAGCCTACACCATCATCCGCCGCGCAGGCGGTATCGGCGAAGGC AACGGCCCCTGGGTCTCCCTCCACGACGGTTTCTTCGGCCGCGACCGCTGGTCCGGCGTCTTCCCCAACGCCGAC CGCCTCGCCCTCGACGTCCATCCATACCTCGGCTTCGGCTCTCAATCCTCCGCCCCCATGTCCTCCTACGCCGAC ACGCCCTGTCGCGCCTGGGGCCGCCTGGTGAACAACTCGATGGCGGCGTTTGGGTTGACGACGGCGGGCGAGTGG AGTAATGCAGTGAATGATTGTGGGTTGTATTTGAATGGGGTGAATTTGGGGACGAGGTATGAGGGGACGTATACC CCTGGGTCGTGGCCGCGTGTGGGTGATTGTAGGGATTGGACGGATTGGACGAGGTATACGCCCGAGTTGAAGAGG GAGATTAGGCAGTTTACGTTGGCGTCTATGGATGCTCTCCAGCACTTCTTCTTTTGGACATGGCGTATCGGCGAT TCGCTCGCCAGTGGACGCGTCGAAACTCCGGCATGGTCTTATGTCCTCGGTTTGCAAGAAGGCTGGATGCCAACC GACCCGCGCGACGCAGACGGTGTATGCAACAACATCGCGCCCTGGACACCCCCTCTCTCCGTCGGCTCCGGTGCG ATCCCCGCCTCCGTCACCTCCGCCCTGTCCTGGCCTCCTCCAGCGATCTCCAACGGCGGCCCCATCGAGGTCCTG CCTTCCTACACCCAAACCGGTGCTATCCCCACTTTGACTGCCACCCCCGTGGAGGCTGCGCCTGGTGAGACGATT ACGCGTACGGTGGATGTGGGTAGTGGGTGGAATAATCCGGAGGATAGTGAGGGGTATGTTGCTGAGGTTGAGGGG TGTAGTTATTTGGATCCGTGGGTTACGCCTGCTGCTGCGCCGCCTTCACCGCTTTGTGTTGGTGTGGCGAGGAGG GGGATGTTGGGGAGGGGTGTTGTGGAGGCTAGGGCTACGCCTGCTCCGTAA |
| Length | 2301 |
Transcript
| Sequence id | CopciAB_500464.T0 |
|---|---|
| Sequence |
>CopciAB_500464.T0 AGACGACAACACGTCGCTAACACACGGCCCCTGCTTCCCTTCCTCATCCCTTCCTTTCGTCTGTGACCCAGTATA ATAGTCCTCGTTGACTAGGGGCTCTCTCTTCCTCTCCTTTCTTTTATAACCTTTTTTCTCTTTTTTGGTTCGTGT TCCGCAAAGCACGTACACTCGACCTCGCATCGTTTGAGGACGTGACAGGTGTCTTTGTCTTGTGCGGACTCGAAC ATGTCGCATAACCCCCAGCAATCTGTAACGGGCGGGGGCGCCTACGACCCGCTCCCCCTCACCGGCGGCGAAGGA CACCAACCGCGGTTTATGGGCCAATACGACGCTGCCGCCTCGTCGTCGCAGCAGAGCCTGCAAGCGCCATCCGAG TATGGCTCGGCCCACCGCCTCAACGACGCCAACGATGTCAATGCAGCACCTTACCGCGACGACCCGAATGCCGTG CCCATGTCGCCGTTGGGTGCGTCTGCTGCCGCTATGAACGAGAAGTCCGGAGGAGGAGGGGGGAGTGGCGGCGAG GGTGGACGGTCGCGCCGCAAGATGGTGATCATCGGCGCCATCGTCGCGCTCATCATCATCATCCTAGCCATTATC CTCCCCGTCTACTTCACCGTGATCAAGAAGGACGATAATGATAGTGCTGGTGGTGGCGACCGGGATAGCAGTAAC GCGGACGAGGGTAAGGATGGGAGTGGGACGGCTAGGCCGAGTGGGAGTGGGAGTCCTGAGCCGGACGTTGAGCTG CCCCCTGTTACTGGCGGCGATGGGAGTATCATTACGATGGAGGATGGTACGACTTTTACGTATCGCAATCCGTTT GGAGGGTATTGGTATTATGATCCCAATGATCCTTTTAATAATGGGGCGAGGGCGCAGTCGTGGACTCCGGCGTTG AATGAGACGTTTAACTATGGTGTTGATAGGATTCGGGGTGTTAATGTTGGTGGTTGGCTTGTTACTGAGCCATTC ATTTCACCGGCTTTGTATGAGCGATATCTCAACGACCCTGCTGGACCTGCCATCGATGAATACGACCTCAGCTTG CGCATGCGCGCTGATACAGCCAACGGAGGCATCAACCAACTCGAGGAACATTATAGGACTTTCATCACTGAGAAG GACTTTGCTGATATCGCTGGTGCCGGTCTGAACTATGTCCGTATTCCTATTGGTTGGTGGGCCGTCGAGACGCGT GGCGATGAGCCTCACTTGGAGGGTGTTTCTTGGAACTACTTCCTCAAGGCGATCAAGTGGGCTAGGAAATACGGT CTCAGGATCAACCTCGATCTTCACGCTGTACCCGGAAGCCAGAACGCCTGGAACCACTCGGGCAAGTTTGGCAGC ATCGGATTCTTGCACGGTCCTATGGGTTACGCCAATGCGCAGAGGACGCTCGATATTATCCGTGTCCTTGCCGAG TTCATCTCGCAGCCTCAGTATCGGGACGTGGTCACCATGTTTGGTATCCTCAATGAGCCTTTGGGTGATCCTATG GGCTTTGATGCTCTAGCTCGATTCTACATGGAAGCCTACACCATCATCCGCCGCGCAGGCGGTATCGGCGAAGGC AACGGCCCCTGGGTCTCCCTCCACGACGGTTTCTTCGGCCGCGACCGCTGGTCCGGCGTCTTCCCCAACGCCGAC CGCCTCGCCCTCGACGTCCATCCATACCTCGGCTTCGGCTCTCAATCCTCCGCCCCCATGTCCTCCTACGCCGAC ACGCCCTGTCGCGCCTGGGGCCGCCTGGTGAACAACTCGATGGCGGCGTTTGGGTTGACGACGGCGGGCGAGTGG AGTAATGCAGTGAATGATTGTGGGTTGTATTTGAATGGGGTGAATTTGGGGACGAGGTATGAGGGGACGTATACC CCTGGGTCGTGGCCGCGTGTGGGTGATTGTAGGGATTGGACGGATTGGACGAGGTATACGCCCGAGTTGAAGAGG GAGATTAGGCAGTTTACGTTGGCGTCTATGGATGCTCTCCAGCACTTCTTCTTTTGGACATGGCGTATCGGCGAT TCGCTCGCCAGTGGACGCGTCGAAACTCCGGCATGGTCTTATGTCCTCGGTTTGCAAGAAGGCTGGATGCCAACC GACCCGCGCGACGCAGACGGTGTATGCAACAACATCGCGCCCTGGACACCCCCTCTCTCCGTCGGCTCCGGTGCG ATCCCCGCCTCCGTCACCTCCGCCCTGTCCTGGCCTCCTCCAGCGATCTCCAACGGCGGCCCCATCGAGGTCCTG CCTTCCTACACCCAAACCGGTGCTATCCCCACTTTGACTGCCACCCCCGTGGAGGCTGCGCCTGGTGAGACGATT ACGCGTACGGTGGATGTGGGTAGTGGGTGGAATAATCCGGAGGATAGTGAGGGGTATGTTGCTGAGGTTGAGGGG TGTAGTTATTTGGATCCGTGGGTTACGCCTGCTGCTGCGCCGCCTTCACCGCTTTGTGTTGGTGTGGCGAGGAGG GGGATGTTGGGGAGGGGTGTTGTGGAGGCTAGGGCTACGCCTGCTCCGTAAGGAGGGTTGTTCCTGTATATGTGC TATACCTGCTTCGTAGAGGTGGGAGTTGTTTTGGTGAAGGAGGATGGGGTGTGTGTTGTGATATGTGATCTAGTG TGGATGACGGCACTATCCGCTGTCTTGCTTCCTGTTTTACTCGAGTCTCTCGTTTATTTTTTGGTCTTGCTTCGG TTTTTGGATTCCCAAGTACCTTACGCCATCTTTCTATACGCATGGACGTTCTCGTTTATTTTTCCCTCCTGGACT TGCTCTGTGTATACTTAACAATACAGATACGCAGTACATATCTGCTTATTTACTAACCGACCCTATTACTTACTT GTTGGGTGGTTTTAGTTATTGGCTGATCTACTTTTGGCCGATCTAGTTTGCTGACTGCAGTCTGTCGTTTACGTG CATTTTTTCCCTCGTTCTATTTCCTTAGTCGTTGCGCGCTTTGCGCGATTCAGAGATCTACGTACATGTATAGCG TCGTTAAGCTTCGACAATTTAGGAGCGTCCCTGAA |
| Length | 3035 |
Gene
| Sequence id | CopciAB_500464.T0 |
|---|---|
| Sequence |
>CopciAB_500464.T0 AGACGACAACACGTCGCTAACACACGGCCCCTGCTTCCCTTCCTCATCCCTTCCTTTCGTCTGTGACCCAGTATA ATAGTCCTCGTTGACTAGGGGCTCTCTCTTCCTCTCCTTTCTTTTATAACCTTTTTTCTCTTTTTTGGTTCGTGT TCCGCAAAGCACGTACACTCGACCTCGCATCGTTTGAGGACGTGACAGGTGTCTTTGTCTTGTGCGGACTCGAAC ATGTCGCATAACCCCCAGCAATCTGTAACGGGCGGGGGCGCCTACGACCCGCTCCCCCTCACCGGCGGCGAAGGA CACCAACCGCGGTTTATGGGCCAATACGACGCTGCCGCCTCGTCGTCGCAGCAGAGCCTGCAAGCGCCATCCGAG TATGGCTCGGCCCACCGCCTCAACGACGCCAACGATGTCAATGCAGCACCTTACCGCGACGACCCGAATGCCGTG CCCATGTCGCCGTTGGGTGCGTCTGCTGCCGCTATGAACGAGAAGTCCGGAGGAGGAGGGGGGAGTGGCGGCGAG GGTGGACGGTCGCGCCGCAAGATGGTGATCATCGGCGCCATCGTCGCGCTCATCATCATCATCCTAGCCATTATC CTCCCCGTCTACTTCACCGTGATCAAGAAGGACGATAATGATAGTGCTGGTGGTGGCGACCGGGATAGCAGTAAC GCGGACGAGGGTAAGGATGGGAGTGGGACGGCTAGGCCGAGTGGGAGTGGGAGTCCTGAGCCGGACGTTGAGCTG CCCCCTGTTACTGGCGGCGATGGGAGTATCATTACGATGGAGGATGGTACGACTTTTACGTATCGCAATCCGTTT GGAGGGTATTGGTATTATGATCCCAATGATCCTTTTAATAATGGGGCGAGGGCGCAGTCGTGGACTCCGGCGTTG AATGAGACGTTTAACTATGGTGTTGATAGGATTCGGGGGTGCGTGTTCCTGGTCTTTTTTCGTGTGGGGGATTTG AGCTGATGGACTTGGATGTAGTGTTAATGTTGGTGGTTGGCTTGTTACTGAGCCATTCATGTGAGTACGGTTCTT GTGGTTGAAGGGTGGTTTCTCTGATGGTTTTGTTTCTAGTTCACCGGCTTTGTATGAGCGATATCTCAACGACCC TGCTGGACCTGCCATCGATGAATACGACCTCAGCTTGCGCATGCGCGCTGATACAGCCAACGGAGGCATCAACCA ACTCGAGGAACATTATAGGACTTTCATCGTATGTTTCTCTTCCTTTCCTCCCCCTTTGGAACCCCCGCTAATGAT TGGGATTCGGGATAGACTGAGAAGGACTTTGCTGATATCGCTGGTGCCGGTCTGAACTATGTCCGTATTCCTATT GGTTGGTGGGCCGTCGAGACGCGTGGCGATGAGCCTCACTTGGAGGGTGTTTCTTGGAAGTCAGTCGCTCTTTAA CCTCTATCCCTTTCGCCCTTCTCATTGACGTTCCGTTTTCTTTGTCAAACAGCTACTTCCTCAAGGCGATCAAGT GGGCTAGGAAATACGGTCTCAGGATCAACCTCGATCTTCACGCTGTACCCGGAAGCCAGAACGCCTGGAACCACT CGGGCAAGTTTGGCAGCATCGGATTCTTGCACGGTCCTATGGGTTACGCCAATGCGCAGAGGACGCTCGATATTA TCCGTGTCCTTGCCGAGTTCATCTCGCAGCCTCAGTATCGGGACGTGGTCACCATGTTTGGTATCCTCAATGAGC CTTTGGGTGATCCTATGGGCTTTGATGCTCTAGCTCGATTGTGAGTTGGGCCTTCTCTTTGCACAATGTACAGTT GTCTCATAGGTTTCCTCTAGCTACATGGAAGCCTACACCATCATCCGCCGCGCAGGCGGTATCGGCGAAGGCAAC GGCCCCTGGGTCTCCCTCCACGACGGTTTCTTCGGCCGCGACCGCTGGTCCGGCGTCTTCCCCAACGCCGACCGC CTCGCCCTCGACGTCCATCCATACCTCGGCTTCGGCTCTCAATCCTCCGCCCCCATGTCCTCCTACGCCGACACG CCCTGTCGCGCCTGGGGCCGCCTGGTGAACAACTCGATGGCGGCGTTTGGGTTGACGACGGCGGGCGAGTGGAGT AATGCAGTGAATGATTGTGGGTTGTATTTGAATGGGGTGAATTTGGGGACGAGGTATGAGGGGACGTATACCCCT GGGTCGTGGCCGCGTGTGGGTGATTGTAGGGATTGGACGGATTGGACGAGGTATACGCCCGAGTTGAAGAGGGAG ATTAGGCAGTTTACGTTGGCGTCTATGGATGCTCTCCAGGTGCGCTTCCTCCCCCATTCCTCTTCTCGAACTGGC TCCATATTCTAACGCGTTTTTTTCTACTCCCTATAGCACTTCTTCTTTTGGACATGGCGTATCGGCGATTCGCTC GCCAGTGGACGCGTCGAAACTCCGGCATGGTCTTATGTCCTCGGTTTGCAAGAAGGCTGGATGCCAACCGACCCG CGCGACGCAGACGGTGTATGCAACAACATCGCGCCCTGGACACCCCCTCTCTCCGTCGGCTCCGGTGCGATCCCC GCCTCCGTCACCTCCGCCCTGTCCTGGCCTCCTCCAGCGATCTCCAACGGCGGCCCCATCGAGGTCCTGCCTTCC TACACCCAAACCGGTGCTATCCCCACTTTGACTGCCACCCCCGTGGAGGCTGCGCCTGGTGAGACGATTACGCGT ACGGTGGATGTGGGTAGTGGGTGGAATAATCCGGAGGATAGTGAGGGGTATGTTGCTGAGGTTGAGGGGTGTAGT TATTTGGATCCGTGGGTTACGCCTGCTGCTGCGCCGCCTTCACCGCTTTGTGTTGGTGTGGCGAGGAGGGGGATG TTGGGGAGGGGTGTTGTGGAGGCTAGGGCTACGCCTGCTCCGTAAGGAGGGTTGTTCCTGTATATGTGCTATACC TGCTTCGTAGAGGTGGGAGTTGTTTTGGTGAAGGAGGATGGGGTGTGTGTTGTGATATGTGATCTAGTGTGGATG ACGGCACTATCCGCTGTCTTGCTTCCTGTTTTACTCGAGTCTCTCGTTTATTTTTTGGTCTTGCTTCGGTTTTTG GATTCCCAAGTACCTTACGCCATCTTTCTATACGCATGGACGTTCTCGTTTATTTTTCCCTCCTGGACTTGCTCT GTGTATACTTAACAATACAGATACGCAGTACATATCTGCTTATTTACTAACCGACCCTATTACTTACTTGTTGGG TGGTTTTAGTTATTGGCTGATCTACTTTTGGCCGATCTAGTTTGCTGACTGCAGTCTGTCGTTTACGTGCATTTT TTCCCTCGTTCTATTTCCTTAGTCGTTGCGCGCTTTGCGCGATTCAGAGATCTACGTACATGTATAGCGTCGTTA AGCTTCGACAATTTAGGAGCGTCCCTGAA |
| Length | 3404 |