CopciAB_500579
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_500579 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 500579 |
| Uniprot id | Functional description | Protein kinase subdomain-containing protein PKL ccin9 | |
| Location | scaffold_4:646716..648651 | Strand | + |
| Gene length (nt) | 1936 | Transcript length (nt) | 1829 |
| CDS length (nt) | 1752 | Protein length (aa) | 583 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7270391 | 40.6 | 1.737E-104 | 351 |
| Flammulina velutipes | Flave_chr10AA00840 | 35.8 | 3.871E-13 | 72 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_500579.T0 |
| Description | Protein kinase subdomain-containing protein PKL ccin9 |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF20147 | Crinkler effector protein N-terminal domain | IPR045379 | 5 | 97 |
| Pfam | PF06293 | Lipopolysaccharide kinase (Kdo/WaaP) family | - | 488 | 537 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR045379 | Crinkler effector protein, N-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Protein kinase subdomain-containing protein PKL ccin9 |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
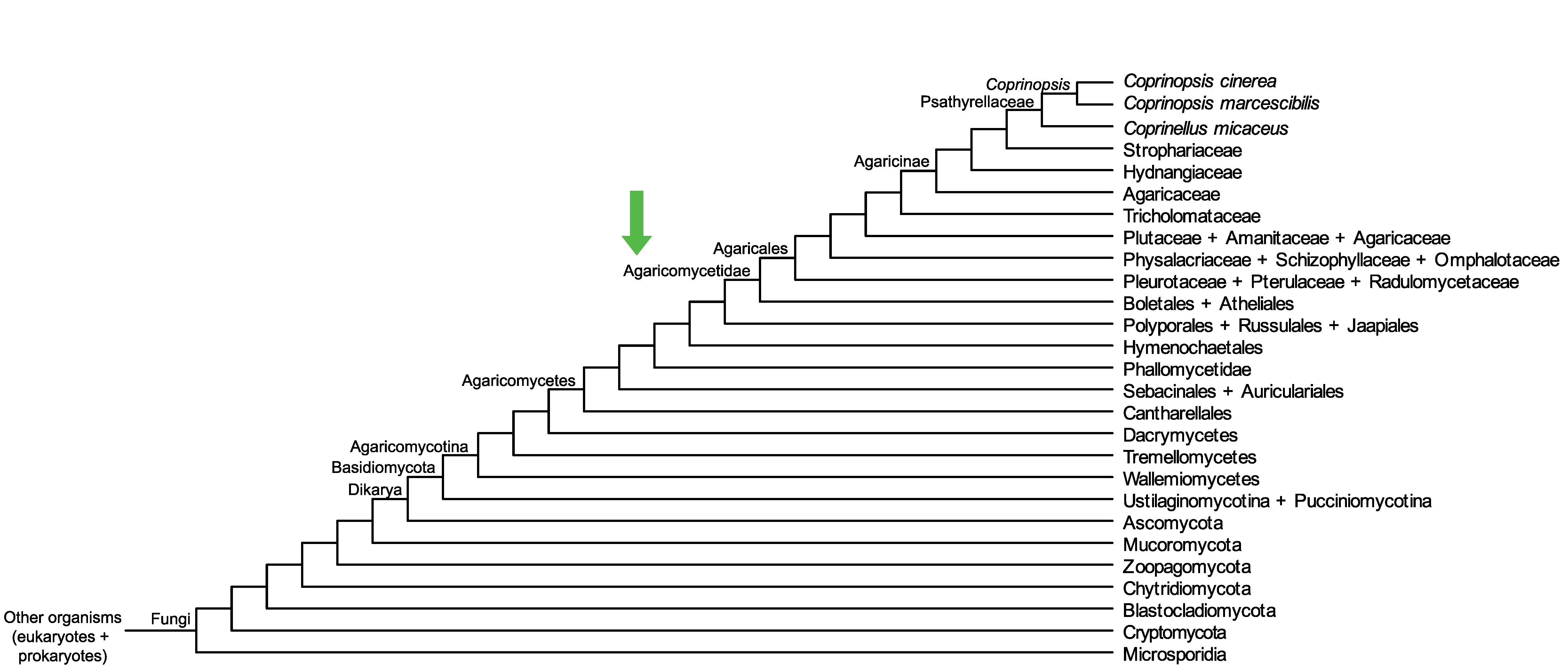
Conservation of CopciAB_500579 across fungi.
Arrow shows the origin of gene family containing CopciAB_500579.
Protein
| Sequence id | CopciAB_500579.T0 |
|---|---|
| Sequence |
>CopciAB_500579.T0 MAETISINCLIYGDGPDRAFTVEIQASDNISILKQLIATGRRLSSIDPTDIHLFKVSLTPDELHRTIPNPKDELK SPLALISTIFKDLSRDKVHVFAVLPDTGLKRSAPESVFQLDALKRARVVTESPSQLAHKWIYESLQRQSTQRIFD DRPEPDSEIPPLALLYQGFGMFMDDFLKRIHVKNFLSTVPDFVDVKDFMGAVDQFAHAMSRFYKDEAERRDVGLY HLNAIMTSTGVDHPPRLYAAFIGKVISDGHAVTEDGFPIHVVQFKNEMVGVTSHAGTEAVGYVARLHNTVREERP TVLQPWRVPCIVTTIVGHEVRFYGVILLGHQYRQISLTPALSCLQGASDGAERMSLYAAFFAAVRLRDRILADLQ QHLDNTHVDPIPPNAILFPAVSKLAKYDGVGDFEFKIIDYRYPRTDCRLLYIALAADDSAVLIKFSRTYCRELHE YCYNKGYAPKLLGFQQLPGGWYGIAMEYDPEMVPIHTENDRALRKPIEQLMKGFHEEGWVHGDLRYCNILRSEDK QKVWVVDFDWGGREGEVCYPTPRLNELLVEGRNCSHWKIRREDDERILDLTFTRLGQV |
| Length | 583 |
Coding
| Sequence id | CopciAB_500579.T0 |
|---|---|
| Sequence |
>CopciAB_500579.T0 ATGGCGGAGACCATCTCGATTAATTGCCTTATTTATGGCGATGGTCCCGACAGGGCGTTTACTGTAGAGATCCAG GCGAGCGATAACATCAGCATCCTCAAACAATTGATAGCGACGGGCAGGCGACTGTCGAGTATCGATCCAACGGAC ATACATCTCTTCAAGGTTTCCCTGACGCCAGACGAGCTACACAGAACGATTCCAAACCCAAAGGACGAGCTCAAA TCACCGCTTGCATTGATATCAACCATCTTCAAAGACTTATCGAGGGACAAAGTCCATGTTTTCGCAGTCCTCCCT GATACTGGCCTCAAACGATCTGCTCCCGAGTCTGTGTTCCAATTAGACGCACTCAAGCGGGCCAGGGTGGTGACT GAATCCCCGTCACAGCTTGCCCATAAGTGGATTTATGAATCGCTACAACGCCAATCTACCCAGAGGATCTTTGAC GACCGTCCTGAACCCGACTCTGAAATTCCCCCTCTTGCGCTCCTATACCAGGGCTTCGGAATGTTCATGGATGAC TTCCTCAAACGCATTCACGTCAAGAACTTCCTGAGCACTGTCCCAGACTTCGTCGATGTCAAGGACTTCATGGGT GCTGTCGATCAATTCGCGCATGCTATGTCCCGGTTCTACAAGGATGAAGCAGAGAGGCGTGATGTTGGACTATAT CATCTCAATGCCATCATGACCTCAACCGGTGTTGACCATCCTCCTCGACTCTATGCGGCCTTCATTGGGAAAGTC ATCAGTGATGGACATGCGGTCACCGAGGACGGTTTCCCCATCCACGTTGTACAATTCAAGAATGAAATGGTTGGG GTCACCTCCCATGCTGGCACTGAAGCCGTCGGCTATGTGGCGCGCCTTCATAACACTGTAAGAGAGGAACGTCCG ACTGTTTTGCAGCCGTGGAGGGTGCCTTGCATTGTTACCACAATTGTTGGACACGAAGTCCGGTTCTACGGCGTC ATCTTGCTCGGACACCAGTACCGCCAGATCAGCCTTACACCAGCTCTGTCATGTCTCCAAGGTGCCAGCGACGGG GCGGAACGAATGTCCCTATACGCCGCGTTCTTCGCCGCCGTCCGACTCCGAGACCGCATCCTCGCAGATCTCCAA CAACACCTCGACAACACGCACGTCGATCCCATCCCACCTAATGCAATTCTCTTCCCAGCCGTTTCAAAGCTCGCC AAATACGACGGTGTTGGCGACTTTGAATTCAAGATCATAGACTACCGATATCCTCGAACAGACTGTCGGCTCCTC TATATCGCCCTTGCCGCTGATGACTCCGCTGTCCTCATCAAGTTCTCTAGGACCTACTGTCGTGAGCTGCATGAA TACTGCTACAACAAGGGCTATGCCCCTAAACTGCTTGGCTTCCAACAACTCCCTGGTGGGTGGTATGGAATCGCG ATGGAGTACGATCCGGAGATGGTCCCCATCCACACCGAAAATGACCGTGCGCTTAGGAAGCCGATTGAGCAACTG ATGAAAGGGTTCCACGAAGAAGGGTGGGTGCATGGAGACCTCCGATATTGCAACATCCTCAGGAGCGAAGACAAG CAAAAGGTTTGGGTTGTCGACTTTGATTGGGGAGGGAGAGAGGGTGAAGTCTGCTATCCGACTCCGAGGCTGAAT GAATTGTTGGTTGAAGGGAGGAACTGTTCTCATTGGAAGATTCGGAGGGAGGACGATGAGAGGATTTTGGATTTG ACGTTCACGAGGCTCGGTCAAGTGTAG |
| Length | 1752 |
Transcript
| Sequence id | CopciAB_500579.T0 |
|---|---|
| Sequence |
>CopciAB_500579.T0 GCTTTGTTGCTTTCCCAACCATGGCGGAGACCATCTCGATTAATTGCCTTATTTATGGCGATGGTCCCGACAGGG CGTTTACTGTAGAGATCCAGGCGAGCGATAACATCAGCATCCTCAAACAATTGATAGCGACGGGCAGGCGACTGT CGAGTATCGATCCAACGGACATACATCTCTTCAAGGTTTCCCTGACGCCAGACGAGCTACACAGAACGATTCCAA ACCCAAAGGACGAGCTCAAATCACCGCTTGCATTGATATCAACCATCTTCAAAGACTTATCGAGGGACAAAGTCC ATGTTTTCGCAGTCCTCCCTGATACTGGCCTCAAACGATCTGCTCCCGAGTCTGTGTTCCAATTAGACGCACTCA AGCGGGCCAGGGTGGTGACTGAATCCCCGTCACAGCTTGCCCATAAGTGGATTTATGAATCGCTACAACGCCAAT CTACCCAGAGGATCTTTGACGACCGTCCTGAACCCGACTCTGAAATTCCCCCTCTTGCGCTCCTATACCAGGGCT TCGGAATGTTCATGGATGACTTCCTCAAACGCATTCACGTCAAGAACTTCCTGAGCACTGTCCCAGACTTCGTCG ATGTCAAGGACTTCATGGGTGCTGTCGATCAATTCGCGCATGCTATGTCCCGGTTCTACAAGGATGAAGCAGAGA GGCGTGATGTTGGACTATATCATCTCAATGCCATCATGACCTCAACCGGTGTTGACCATCCTCCTCGACTCTATG CGGCCTTCATTGGGAAAGTCATCAGTGATGGACATGCGGTCACCGAGGACGGTTTCCCCATCCACGTTGTACAAT TCAAGAATGAAATGGTTGGGGTCACCTCCCATGCTGGCACTGAAGCCGTCGGCTATGTGGCGCGCCTTCATAACA CTGTAAGAGAGGAACGTCCGACTGTTTTGCAGCCGTGGAGGGTGCCTTGCATTGTTACCACAATTGTTGGACACG AAGTCCGGTTCTACGGCGTCATCTTGCTCGGACACCAGTACCGCCAGATCAGCCTTACACCAGCTCTGTCATGTC TCCAAGGTGCCAGCGACGGGGCGGAACGAATGTCCCTATACGCCGCGTTCTTCGCCGCCGTCCGACTCCGAGACC GCATCCTCGCAGATCTCCAACAACACCTCGACAACACGCACGTCGATCCCATCCCACCTAATGCAATTCTCTTCC CAGCCGTTTCAAAGCTCGCCAAATACGACGGTGTTGGCGACTTTGAATTCAAGATCATAGACTACCGATATCCTC GAACAGACTGTCGGCTCCTCTATATCGCCCTTGCCGCTGATGACTCCGCTGTCCTCATCAAGTTCTCTAGGACCT ACTGTCGTGAGCTGCATGAATACTGCTACAACAAGGGCTATGCCCCTAAACTGCTTGGCTTCCAACAACTCCCTG GTGGGTGGTATGGAATCGCGATGGAGTACGATCCGGAGATGGTCCCCATCCACACCGAAAATGACCGTGCGCTTA GGAAGCCGATTGAGCAACTGATGAAAGGGTTCCACGAAGAAGGGTGGGTGCATGGAGACCTCCGATATTGCAACA TCCTCAGGAGCGAAGACAAGCAAAAGGTTTGGGTTGTCGACTTTGATTGGGGAGGGAGAGAGGGTGAAGTCTGCT ATCCGACTCCGAGGCTGAATGAATTGTTGGTTGAAGGGAGGAACTGTTCTCATTGGAAGATTCGGAGGGAGGACG ATGAGAGGATTTTGGATTTGACGTTCACGAGGCTCGGTCAAGTGTAGATTGTATGCTGAGGTGTATGGAATGACA TGCAACCTGCATACTTGGTTCTTACTCTC |
| Length | 1829 |
Gene
| Sequence id | CopciAB_500579.T0 |
|---|---|
| Sequence |
>CopciAB_500579.T0 GCTTTGTTGCTTTCCCAACCATGGCGGAGACCATCTCGATTAATTGCCTTATTTATGGCGATGGTCCCGACAGGG CGTTTACTGTAGAGATCCAGGCGAGCGATAACATCAGCATCCTCAAACAATTGATAGCGACGGGCAGGCGACTGT CGAGTATCGATCCAACGGACATACATCTCTTCAAGGTTTCCCTGACGCCAGACGAGCTACACAGAACGATTCCAA ACCCAAAGGACGAGCTCAAATCACCGCTTGCATTGATATCAACCATCTTCAAAGACTTATCGAGGGACAAAGTCC ATGTTTTCGCAGGTAAGCAAAGGCTATGACCGTGCTTCATATCTTCACTTAGATTGCTGCAGTCCTCCCTGATAC TGGCCTCAAACGATCTGCTCCCGAGTCTGTGTTCCAATTAGACGCACTCAAGCGGGCCAGGGTGGTGACTGAATC CCCGTCACAGCTTGCCCATAAGTGGATTTATGAATCGCTACAACGCCAATCTACCCAGAGGATCTTTGACGACCG TCCTGAACCCGACTCTGAAATTCCCCCTCTTGCGCTCCTATACCAGGGCTTCGGAATGTTCATGGATGACTTCCT CAAACGCATTCACGTCAAGAACTTCCTGAGCACTGTCCCAGACTTCGTCGATGTCAAGGACTTCATGGGTGCTGT CGATCAATTCGCGCATGCTATGTCCCGGTTCTACAAGGATGAAGCAGAGAGGCGTGATGTTGGACTATATCATCT CAATGCCATCATGACCTCAACCGGTGTTGACCATCCTCCTCGACTCTATGCGGCCTTCATTGGGAAAGTCATCAG TGATGGACATGCGGTCACCGAGGACGGTTTCCCCATCCACGTTGTACAATTCAAGAATGAAATGGTTGGGGTCAC CTCCCATGCTGGCACTGAAGCCGTCGGCTATGTGGCGCGCCTTCATAACACTGTAAGAGAGGAACGTCCGACTGT TTTGCAGCCGTGGAGGGTGCCTTGCATTGTTACCACAATTGTTGGTAGGTACAGGAATACCCTCTCTCTTCCTCG TGCCGTGTCGTTCAAGCTCGTCATAGGACACGAAGTCCGGTTCTACGGCGTCATCTTGCTCGGACACCAGTACCG CCAGATCAGCCTTACACCAGCTCTGTCATGTCTCCAAGGTGCCAGCGACGGGGCGGAACGAATGTCCCTATACGC CGCGTTCTTCGCCGCCGTCCGACTCCGAGACCGCATCCTCGCAGATCTCCAACAACACCTCGACAACACGCACGT CGATCCCATCCCACCTAATGCAATTCTCTTCCCAGCCGTTTCAAAGCTCGCCAAATACGACGGTGTTGGCGACTT TGAATTCAAGATCATAGACTACCGATATCCTCGAACAGACTGTCGGCTCCTCTATATCGCCCTTGCCGCTGATGA CTCCGCTGTCCTCATCAAGTTCTCTAGGACCTACTGTCGTGAGCTGCATGAATACTGCTACAACAAGGGCTATGC CCCTAAACTGCTTGGCTTCCAACAACTCCCTGGTGGGTGGTATGGAATCGCGATGGAGTACGATCCGGAGATGGT CCCCATCCACACCGAAAATGACCGTGCGCTTAGGAAGCCGATTGAGCAACTGATGAAAGGGTTCCACGAAGAAGG GTGGGTGCATGGAGACCTCCGATATTGCAACATCCTCAGGAGCGAAGACAAGCAAAAGGTTTGGGTTGTCGACTT TGATTGGGGAGGGAGAGAGGGTGAAGTCTGCTATCCGACTCCGAGGCTGAATGAATTGTTGGTTGAAGGGAGGAA CTGTTCTCATTGGAAGATTCGGAGGGAGGACGATGAGAGGATTTTGGATTTGACGTTCACGAGGCTCGGTCAAGT GTAGATTGTATGCTGAGGTGTATGGAATGACATGCAACCTGCATACTTGGTTCTTACTCTC |
| Length | 1936 |