CopciAB_501525
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_501525 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 501525 |
| Uniprot id | Functional description | Phosphotransferase | |
| Location | scaffold_4:3359123..3360938 | Strand | - |
| Gene length (nt) | 1816 | Transcript length (nt) | 1816 |
| CDS length (nt) | 1398 | Protein length (aa) | 465 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_186284 | 60 | 8.891E-186 | 581 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_79879 | 59.3 | 4.966E-184 | 576 |
| Lentinula edodes NBRC 111202 | Lenedo1_1003918 | 37.8 | 1.851E-74 | 258 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_8692 | 35.9 | 8.813E-74 | 256 |
| Agrocybe aegerita | Agrae_CAA7266098 | 36.9 | 1.765E-56 | 205 |
| Schizophyllum commune H4-8 | Schco3_2632415 | 32.9 | 9.231E-55 | 200 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_501525.T0 |
| Description | Phosphotransferase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF01636 | Phosphotransferase enzyme family | IPR002575 | 2 | 272 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR002575 | Aminoglycoside phosphotransferase |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| H | Phosphotransferase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
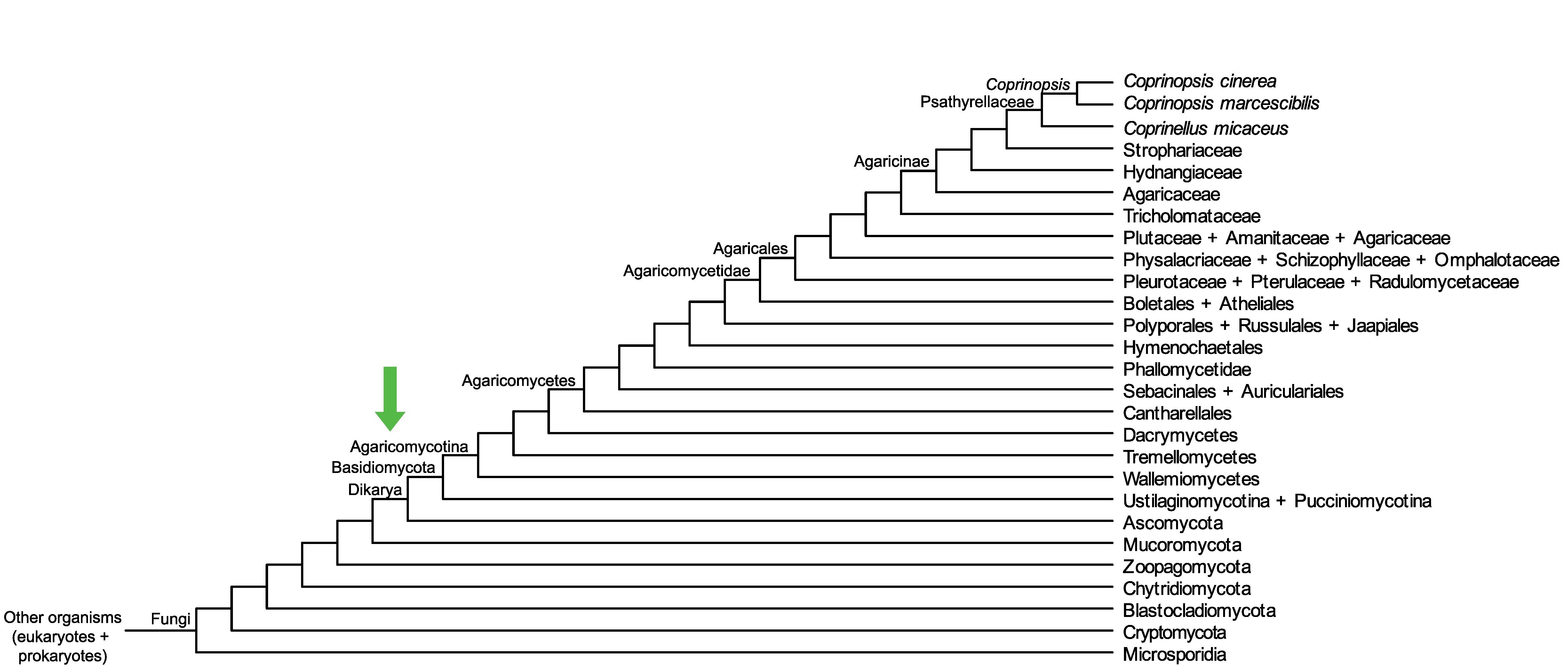
Conservation of CopciAB_501525 across fungi.
Arrow shows the origin of gene family containing CopciAB_501525.
Protein
| Sequence id | CopciAB_501525.T0 |
|---|---|
| Sequence |
>CopciAB_501525.T0 MTDGRRVIARLPTPLAGPPHLVTASEVATMRFLRERLGLTQIPRVLSCSSRAEETPVGAEHIIMDVADGVSLLSV WDELTMSQKLRVVDQWIKFESVATKAISGGYGSLYYRNDLPPELARDLTVWGKVDEEFVLGPSTVQLGFWEGKHG NPRDLELELDRGPWPDVASYLKSISNCERAWIRKFAKPPPSTGRTYRAPWEAPPHLQIPEEHIHLLNLYDNIVAH LIPTDKRYHRPSLTLRDSHHTNIFLSKDALDRDGSIQISSIIDWQHTAVLPLYLTAFAPQFIQQRLFFPGKPKEE VERENAYLHKAYYMLYNDTKLDTVWASLVNPESPGRHMSEQLPGIAQFCWHGGFATLKLDLITAANHWEELVGPG VPFPHPELCKEDVQKAEEDAVQWAEVEKAIEKVRESVGVYEDGWVSNVPEYEEAIEANDEQRKAWVDSLTEEEKK GLGGVDPADIWLLRP |
| Length | 465 |
Coding
| Sequence id | CopciAB_501525.T0 |
|---|---|
| Sequence |
>CopciAB_501525.T0 ATGACCGACGGACGTCGCGTCATCGCTCGACTCCCTACCCCCCTTGCTGGACCTCCACACTTGGTCACCGCAAGC GAAGTCGCCACGATGCGCTTTCTCCGCGAGCGCCTGGGTCTAACACAAATCCCTCGCGTCCTCTCCTGTTCGTCA CGCGCGGAGGAGACGCCAGTCGGAGCAGAGCACATCATCATGGACGTCGCAGACGGCGTGTCGCTCCTCTCCGTC TGGGACGAACTGACCATGTCACAGAAGCTCAGAGTCGTCGACCAGTGGATCAAATTCGAGTCTGTAGCTACCAAA GCCATTAGCGGCGGCTACGGCTCCTTGTACTACCGCAACGACCTTCCACCAGAGCTCGCACGCGACCTCACAGTG TGGGGCAAAGTCGACGAGGAATTCGTACTCGGACCTAGCACCGTGCAGCTCGGCTTCTGGGAAGGCAAGCACGGA AACCCACGCGACCTCGAACTCGAACTTGACCGCGGGCCATGGCCAGACGTTGCCTCCTACCTCAAATCCATCTCC AACTGCGAACGTGCTTGGATCCGCAAATTCGCCAAACCACCACCCTCTACAGGCCGCACCTACAGAGCCCCATGG GAAGCCCCTCCCCATCTCCAAATACCCGAAGAGCACATCCACCTCCTCAACCTCTACGACAATATCGTGGCCCAC CTCATCCCCACAGACAAGCGCTACCACCGCCCCTCTCTCACCCTCCGCGACTCGCATCACACCAACATCTTCCTC TCCAAAGACGCGCTCGACCGCGATGGCTCAATCCAAATCTCCTCCATAATCGACTGGCAACACACCGCCGTCCTC CCTCTCTACCTCACAGCCTTCGCCCCCCAATTCATCCAACAACGCCTCTTCTTCCCCGGAAAACCCAAAGAGGAA GTCGAAAGGGAAAACGCGTACCTCCACAAGGCGTACTACATGCTCTATAACGACACCAAGCTCGATACCGTCTGG GCCTCCCTCGTCAACCCCGAATCTCCAGGACGACACATGTCCGAGCAGTTGCCAGGAATCGCGCAGTTTTGCTGG CACGGCGGGTTTGCGACTCTCAAGTTGGACCTCATCACCGCGGCGAATCACTGGGAGGAGCTTGTAGGACCCGGA GTACCCTTTCCTCATCCAGAGCTCTGCAAGGAGGACGTTCAGAAGGCGGAGGAAGACGCAGTGCAGTGGGCCGAG GTCGAGAAAGCGATCGAAAAGGTTCGGGAATCTGTTGGCGTATATGAGGATGGGTGGGTATCAAATGTGCCAGAG TACGAAGAAGCCATCGAGGCCAACGATGAGCAGCGCAAAGCTTGGGTGGACAGCTTGACTGAGGAGGAAAAGAAA GGACTGGGCGGTGTGGACCCTGCAGATATTTGGCTTCTTCGACCTTGA |
| Length | 1398 |
Transcript
| Sequence id | CopciAB_501525.T0 |
|---|---|
| Sequence |
>CopciAB_501525.T0 CCCTTTCTACTATCCGCCGCCCTGCATCCCCGAATCCTCTCCGCACCCAACCTACGACGCTTTCGTCTCCTCAGT ACGAAAGCGTCTCCAAACGATGACTTCTTCCGCTACACAACCGGCCGATGGCTCCAAAACGAACAACGCCACCAA TCCCTACGATACCAACCCTTCAACATCGAAGCACTAAAGGACGCCGCCACGCGCAGCGCAAACGCAACTGGAGTG GCCGGGTTCAAAAAACTCGCAGAAGGACGATGCAACAAAGTGTTCGATGTACAAATGACCGACGGACGTCGCGTC ATCGCTCGACTCCCTACCCCCCTTGCTGGACCTCCACACTTGGTCACCGCAAGCGAAGTCGCCACGATGCGCTTT CTCCGCGAGCGCCTGGGTCTAACACAAATCCCTCGCGTCCTCTCCTGTTCGTCACGCGCGGAGGAGACGCCAGTC GGAGCAGAGCACATCATCATGGACGTCGCAGACGGCGTGTCGCTCCTCTCCGTCTGGGACGAACTGACCATGTCA CAGAAGCTCAGAGTCGTCGACCAGTGGATCAAATTCGAGTCTGTAGCTACCAAAGCCATTAGCGGCGGCTACGGC TCCTTGTACTACCGCAACGACCTTCCACCAGAGCTCGCACGCGACCTCACAGTGTGGGGCAAAGTCGACGAGGAA TTCGTACTCGGACCTAGCACCGTGCAGCTCGGCTTCTGGGAAGGCAAGCACGGAAACCCACGCGACCTCGAACTC GAACTTGACCGCGGGCCATGGCCAGACGTTGCCTCCTACCTCAAATCCATCTCCAACTGCGAACGTGCTTGGATC CGCAAATTCGCCAAACCACCACCCTCTACAGGCCGCACCTACAGAGCCCCATGGGAAGCCCCTCCCCATCTCCAA ATACCCGAAGAGCACATCCACCTCCTCAACCTCTACGACAATATCGTGGCCCACCTCATCCCCACAGACAAGCGC TACCACCGCCCCTCTCTCACCCTCCGCGACTCGCATCACACCAACATCTTCCTCTCCAAAGACGCGCTCGACCGC GATGGCTCAATCCAAATCTCCTCCATAATCGACTGGCAACACACCGCCGTCCTCCCTCTCTACCTCACAGCCTTC GCCCCCCAATTCATCCAACAACGCCTCTTCTTCCCCGGAAAACCCAAAGAGGAAGTCGAAAGGGAAAACGCGTAC CTCCACAAGGCGTACTACATGCTCTATAACGACACCAAGCTCGATACCGTCTGGGCCTCCCTCGTCAACCCCGAA TCTCCAGGACGACACATGTCCGAGCAGTTGCCAGGAATCGCGCAGTTTTGCTGGCACGGCGGGTTTGCGACTCTC AAGTTGGACCTCATCACCGCGGCGAATCACTGGGAGGAGCTTGTAGGACCCGGAGTACCCTTTCCTCATCCAGAG CTCTGCAAGGAGGACGTTCAGAAGGCGGAGGAAGACGCAGTGCAGTGGGCCGAGGTCGAGAAAGCGATCGAAAAG GTTCGGGAATCTGTTGGCGTATATGAGGATGGGTGGGTATCAAATGTGCCAGAGTACGAAGAAGCCATCGAGGCC AACGATGAGCAGCGCAAAGCTTGGGTGGACAGCTTGACTGAGGAGGAAAAGAAAGGACTGGGCGGTGTGGACCCT GCAGATATTTGGCTTCTTCGACCTTGATAGTAATGGTGTACCCTTGCTTTCACGCATAATTACATGGCCCCTGCT CTTATGGTCAGGCGATAGATTACACCCTAGAAAACACTGGTGGTCGTTGTCATAGCTTCGAAGGAAATACTAAAG GGGTTCACGATACATA |
| Length | 1816 |
Gene
| Sequence id | CopciAB_501525.T0 |
|---|---|
| Sequence |
>CopciAB_501525.T0 CCCTTTCTACTATCCGCCGCCCTGCATCCCCGAATCCTCTCCGCACCCAACCTACGACGCTTTCGTCTCCTCAGT ACGAAAGCGTCTCCAAACGATGACTTCTTCCGCTACACAACCGGCCGATGGCTCCAAAACGAACAACGCCACCAA TCCCTACGATACCAACCCTTCAACATCGAAGCACTAAAGGACGCCGCCACGCGCAGCGCAAACGCAACTGGAGTG GCCGGGTTCAAAAAACTCGCAGAAGGACGATGCAACAAAGTGTTCGATGTACAAATGACCGACGGACGTCGCGTC ATCGCTCGACTCCCTACCCCCCTTGCTGGACCTCCACACTTGGTCACCGCAAGCGAAGTCGCCACGATGCGCTTT CTCCGCGAGCGCCTGGGTCTAACACAAATCCCTCGCGTCCTCTCCTGTTCGTCACGCGCGGAGGAGACGCCAGTC GGAGCAGAGCACATCATCATGGACGTCGCAGACGGCGTGTCGCTCCTCTCCGTCTGGGACGAACTGACCATGTCA CAGAAGCTCAGAGTCGTCGACCAGTGGATCAAATTCGAGTCTGTAGCTACCAAAGCCATTAGCGGCGGCTACGGC TCCTTGTACTACCGCAACGACCTTCCACCAGAGCTCGCACGCGACCTCACAGTGTGGGGCAAAGTCGACGAGGAA TTCGTACTCGGACCTAGCACCGTGCAGCTCGGCTTCTGGGAAGGCAAGCACGGAAACCCACGCGACCTCGAACTC GAACTTGACCGCGGGCCATGGCCAGACGTTGCCTCCTACCTCAAATCCATCTCCAACTGCGAACGTGCTTGGATC CGCAAATTCGCCAAACCACCACCCTCTACAGGCCGCACCTACAGAGCCCCATGGGAAGCCCCTCCCCATCTCCAA ATACCCGAAGAGCACATCCACCTCCTCAACCTCTACGACAATATCGTGGCCCACCTCATCCCCACAGACAAGCGC TACCACCGCCCCTCTCTCACCCTCCGCGACTCGCATCACACCAACATCTTCCTCTCCAAAGACGCGCTCGACCGC GATGGCTCAATCCAAATCTCCTCCATAATCGACTGGCAACACACCGCCGTCCTCCCTCTCTACCTCACAGCCTTC GCCCCCCAATTCATCCAACAACGCCTCTTCTTCCCCGGAAAACCCAAAGAGGAAGTCGAAAGGGAAAACGCGTAC CTCCACAAGGCGTACTACATGCTCTATAACGACACCAAGCTCGATACCGTCTGGGCCTCCCTCGTCAACCCCGAA TCTCCAGGACGACACATGTCCGAGCAGTTGCCAGGAATCGCGCAGTTTTGCTGGCACGGCGGGTTTGCGACTCTC AAGTTGGACCTCATCACCGCGGCGAATCACTGGGAGGAGCTTGTAGGACCCGGAGTACCCTTTCCTCATCCAGAG CTCTGCAAGGAGGACGTTCAGAAGGCGGAGGAAGACGCAGTGCAGTGGGCCGAGGTCGAGAAAGCGATCGAAAAG GTTCGGGAATCTGTTGGCGTATATGAGGATGGGTGGGTATCAAATGTGCCAGAGTACGAAGAAGCCATCGAGGCC AACGATGAGCAGCGCAAAGCTTGGGTGGACAGCTTGACTGAGGAGGAAAAGAAAGGACTGGGCGGTGTGGACCCT GCAGATATTTGGCTTCTTCGACCTTGATAGTAATGGTGTACCCTTGCTTTCACGCATAATTACATGGCCCCTGCT CTTATGGTCAGGCGATAGATTACACCCTAGAAAACACTGGTGGTCGTTGTCATAGCTTCGAAGGAAATACTAAAG GGGTTCACGATACATA |
| Length | 1816 |