CopciAB_501865
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_501865 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 501865 |
| Uniprot id | Functional description | Altered inheritance of mitochondria protein 9, mitochondrial | |
| Location | scaffold_9:1768884..1770902 | Strand | + |
| Gene length (nt) | 2019 | Transcript length (nt) | 1870 |
| CDS length (nt) | 1707 | Protein length (aa) | 568 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16570 | 35.9 | 5.74E-104 | 349 |
| Flammulina velutipes | Flave_chr10AA01338 | 37.4 | 1.103E-101 | 342 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_80256 | 33.6 | 1.661E-89 | 306 |
| Grifola frondosa | Grifr_OBZ65402 | 31.2 | 6.705E-77 | 269 |
| Agrocybe aegerita | Agrae_CAA7265732 | 34.6 | 8.668E-72 | 254 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1449360 | 28.8 | 1.876E-53 | 199 |
| Lentinula edodes B17 | Lened_B_1_1_3280 | 40.5 | 5.106E-24 | 107 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_501865.T0 |
| Description | Altered inheritance of mitochondria protein 9, mitochondrial |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| No records | |||||
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Altered inheritance of mitochondria protein 9, mitochondrial |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
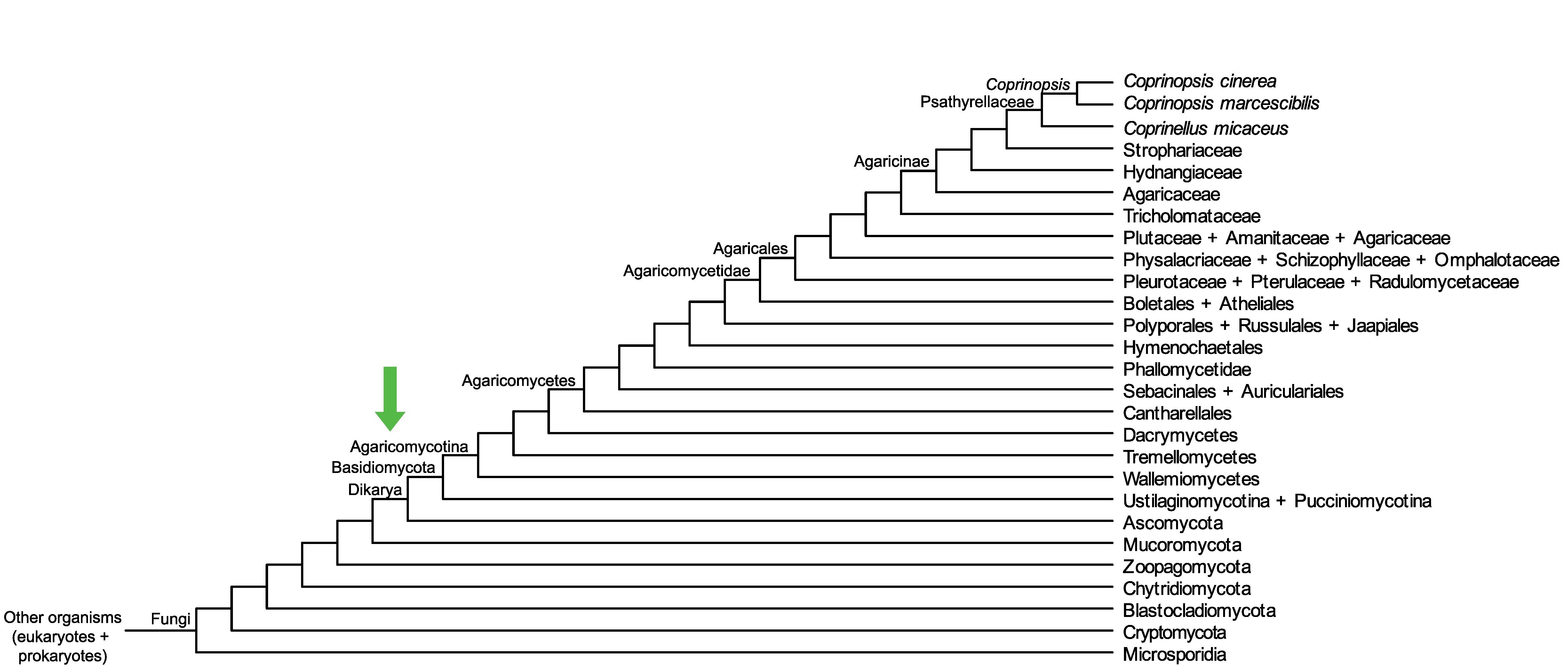
Conservation of CopciAB_501865 across fungi.
Arrow shows the origin of gene family containing CopciAB_501865.
Protein
| Sequence id | CopciAB_501865.T0 |
|---|---|
| Sequence |
>CopciAB_501865.T0 MQRLLLKRLSSGPIQRRLVTSSCLQNPLFSYSTGRWLYNEKDQLAVRHVPFNVDALKEAACSALGKSSCTSFEKI AEGSYNKVFRMYFEDGKTAIARIPSLRLLGNASMITASEVATMEFMRLYRSFHIPLVYSWSKDLDGPVKSPYIIM EDIEGVPLLHDWYKIRGDPVRHAMRRFVTDVRQMLCIPFDQIGGLYFKEDLPPHLQERPLFSPALYQTETALKMF EPAIDRFRVGPIPDYTWWRTMHDEVGADRGPWKDIGDMLIAAVNLEKSAIAKYRRNPSILKKTETTSLEELDVLE GLLEKVVTLAPSLALALSNIPDFINFNVAVHADLGPNNAMVPADNGTNTLSRFDQFTYIDWQGTSILPFPFHCHI PELVQYNGDIIKVPDEYINQIPIPEGLDQLPEDQREYIMAHHRLATRELAFKNAMLDNFHPWAAANAHCLFPYIT QLPDRVLRGVTHGPILLRECLMRISSEWDEEQCDGPCPISFTEDEIAAHDRAAVFRHLYFVNTKTAQKRIDMDGD GWVPNEEYEEARARFEKYRESWDPKVCGGPWPFQDGGYSYFLA |
| Length | 568 |
Coding
| Sequence id | CopciAB_501865.T0 |
|---|---|
| Sequence |
>CopciAB_501865.T0 ATGCAAAGGCTGCTGCTGAAGCGACTAAGTTCTGGCCCTATCCAGAGGCGTTTGGTGACATCTTCATGTCTTCAA AACCCGTTGTTTAGCTACTCTACTGGACGCTGGCTCTACAACGAGAAAGACCAATTGGCTGTCAGACATGTCCCT TTCAACGTCGATGCTCTAAAAGAGGCAGCTTGCAGCGCCCTGGGCAAATCATCATGCACGAGTTTTGAAAAGATT GCGGAGGGCTCATACAACAAAGTCTTCCGAATGTATTTTGAGGATGGAAAAACGGCTATAGCCCGAATTCCTTCA CTCCGTCTGCTCGGGAACGCCTCAATGATCACCGCCAGCGAGGTAGCCACGATGGAGTTCATGAGGCTTTACCGA AGCTTCCACATCCCCCTCGTCTATTCCTGGTCCAAAGACCTCGATGGCCCGGTCAAATCACCCTACATCATCATG GAAGACATTGAAGGTGTCCCTCTCCTCCACGACTGGTACAAAATCCGCGGGGACCCCGTCCGACATGCAATGCGG CGATTCGTCACTGATGTCAGGCAAATGTTATGCATCCCATTCGACCAGATCGGAGGATTGTACTTTAAAGAGGAC CTCCCTCCTCACCTGCAAGAGCGGCCCTTGTTTTCACCCGCACTCTATCAAACCGAGACGGCTCTAAAAATGTTT GAGCCCGCTATTGATAGATTTCGTGTTGGGCCTATTCCGGATTACACCTGGTGGAGGACGATGCATGACGAAGTA GGCGCAGACCGTGGCCCTTGGAAGGATATCGGGGATATGCTCATCGCCGCTGTCAACTTGGAAAAAAGCGCCATT GCCAAATACCGACGTAATCCCTCCATTCTGAAAAAGACAGAGACCACCTCACTTGAAGAGCTTGACGTTCTCGAA GGGCTCCTCGAGAAGGTCGTAACCCTGGCGCCTTCATTGGCTTTGGCCTTGAGCAACATCCCAGATTTCATCAAC TTCAATGTAGCCGTCCATGCTGATTTAGGGCCAAACAACGCTATGGTTCCTGCCGACAACGGCACAAACACACTC AGCCGTTTCGACCAGTTCACATACATCGACTGGCAGGGGACGTCCATCCTCCCCTTCCCCTTCCACTGCCACATA CCCGAGCTCGTGCAATACAACGGCGACATCATCAAAGTCCCAGACGAGTATATAAACCAAATCCCAATTCCAGAA GGACTCGATCAACTTCCAGAAGACCAGCGAGAGTATATCATGGCACACCACCGCCTCGCCACCAGAGAATTGGCC TTTAAAAATGCCATGCTGGACAATTTCCATCCATGGGCCGCTGCGAACGCGCACTGCCTCTTCCCTTATATCACC CAACTCCCCGACCGAGTCCTACGAGGTGTAACACACGGGCCTATCTTGCTTCGAGAATGCCTAATGCGCATATCC TCGGAATGGGACGAGGAGCAGTGTGATGGACCGTGCCCCATTTCTTTCACGGAGGACGAAATTGCCGCGCACGAC CGTGCAGCGGTCTTCCGTCATCTTTACTTTGTCAATACCAAGACTGCGCAAAAGCGTATCGATATGGATGGGGAT GGGTGGGTGCCGAACGAAGAGTACGAGGAGGCGCGGGCGCGGTTTGAGAAGTATCGCGAGTCGTGGGATCCTAAG GTCTGCGGAGGACCCTGGCCTTTCCAAGATGGTGGTTACTCGTACTTCCTAGCCTAA |
| Length | 1707 |
Transcript
| Sequence id | CopciAB_501865.T0 |
|---|---|
| Sequence |
>CopciAB_501865.T0 AGAGAATGCTTTCCGTTGCCCCCTCATCATATGCTTCGCGCTGTCGCCGCTCACTGTGACAGCAGTCGGACCTTG AACCGTGTGACTTGACCTTTGTCTCTGGCTATCGGCGGCTAGGATGCAAAGGCTGCTGCTGAAGCGACTAAGTTC TGGCCCTATCCAGAGGCGTTTGGTGACATCTTCATGTCTTCAAAACCCGTTGTTTAGCTACTCTACTGGACGCTG GCTCTACAACGAGAAAGACCAATTGGCTGTCAGACATGTCCCTTTCAACGTCGATGCTCTAAAAGAGGCAGCTTG CAGCGCCCTGGGCAAATCATCATGCACGAGTTTTGAAAAGATTGCGGAGGGCTCATACAACAAAGTCTTCCGAAT GTATTTTGAGGATGGAAAAACGGCTATAGCCCGAATTCCTTCACTCCGTCTGCTCGGGAACGCCTCAATGATCAC CGCCAGCGAGGTAGCCACGATGGAGTTCATGAGGCTTTACCGAAGCTTCCACATCCCCCTCGTCTATTCCTGGTC CAAAGACCTCGATGGCCCGGTCAAATCACCCTACATCATCATGGAAGACATTGAAGGTGTCCCTCTCCTCCACGA CTGGTACAAAATCCGCGGGGACCCCGTCCGACATGCAATGCGGCGATTCGTCACTGATGTCAGGCAAATGTTATG CATCCCATTCGACCAGATCGGAGGATTGTACTTTAAAGAGGACCTCCCTCCTCACCTGCAAGAGCGGCCCTTGTT TTCACCCGCACTCTATCAAACCGAGACGGCTCTAAAAATGTTTGAGCCCGCTATTGATAGATTTCGTGTTGGGCC TATTCCGGATTACACCTGGTGGAGGACGATGCATGACGAAGTAGGCGCAGACCGTGGCCCTTGGAAGGATATCGG GGATATGCTCATCGCCGCTGTCAACTTGGAAAAAAGCGCCATTGCCAAATACCGACGTAATCCCTCCATTCTGAA AAAGACAGAGACCACCTCACTTGAAGAGCTTGACGTTCTCGAAGGGCTCCTCGAGAAGGTCGTAACCCTGGCGCC TTCATTGGCTTTGGCCTTGAGCAACATCCCAGATTTCATCAACTTCAATGTAGCCGTCCATGCTGATTTAGGGCC AAACAACGCTATGGTTCCTGCCGACAACGGCACAAACACACTCAGCCGTTTCGACCAGTTCACATACATCGACTG GCAGGGGACGTCCATCCTCCCCTTCCCCTTCCACTGCCACATACCCGAGCTCGTGCAATACAACGGCGACATCAT CAAAGTCCCAGACGAGTATATAAACCAAATCCCAATTCCAGAAGGACTCGATCAACTTCCAGAAGACCAGCGAGA GTATATCATGGCACACCACCGCCTCGCCACCAGAGAATTGGCCTTTAAAAATGCCATGCTGGACAATTTCCATCC ATGGGCCGCTGCGAACGCGCACTGCCTCTTCCCTTATATCACCCAACTCCCCGACCGAGTCCTACGAGGTGTAAC ACACGGGCCTATCTTGCTTCGAGAATGCCTAATGCGCATATCCTCGGAATGGGACGAGGAGCAGTGTGATGGACC GTGCCCCATTTCTTTCACGGAGGACGAAATTGCCGCGCACGACCGTGCAGCGGTCTTCCGTCATCTTTACTTTGT CAATACCAAGACTGCGCAAAAGCGTATCGATATGGATGGGGATGGGTGGGTGCCGAACGAAGAGTACGAGGAGGC GCGGGCGCGGTTTGAGAAGTATCGCGAGTCGTGGGATCCTAAGGTCTGCGGAGGACCCTGGCCTTTCCAAGATGG TGGTTACTCGTACTTCCTAGCCTAATACCTTTGTCTATATCTAGTTGTGAAATTTACCTCCTCTCACTAC |
| Length | 1870 |
Gene
| Sequence id | CopciAB_501865.T0 |
|---|---|
| Sequence |
>CopciAB_501865.T0 AGAGAATGCTTTCCGTTGCCCCCTCATCATATGCTTCGCGCTGTCGCCGCTCACTGTGACAGCAGTCGGACCTTG AACCGTGTGACTTGACCTTTGTCTCTGGCTATCGGCGGCTAGGATGCAAAGGCTGCTGCTGAAGCGACTAAGTTC TGGCCCTATCCAGAGGCGTTTGGTGACATCTTCATGTCTTCAAAACCCGTTGTTTAGCTACTCTACTGGACGCTG GCTCTACAACGAGAAAGACCGTGAGTTGAGTAGCTTCATCCAATAGATCCAATGCTGACACTGTCCAGAATTGGC TGTCAGACATGTCCCTTTCAACGTCGATGCTCTAAAAGAGGCAGCTTGCAGCGCCCTGGGCAAATCATCATGCAC GAGTTTTGAAAAGATTGCGGAGGGTGCGGCACAAACTCGAATGGCGCTACGCCGATGCTAATGGTTACGTAGGCT CATACAACAAAGTCTTCCGAATGTATTTTGAGGATGGAAAAACGGCTATAGCCCGAATTCCTTCACTCCGTCTGC TCGGGAACGCCTCAATGATCACCGCCAGCGAGGTAGCCACGATGGAGTTCATGAGGCTTTACCGAAGCTTCCACA TCCCCCTCGTCTATTCCTGGTCCAAAGACCTCGATGGCCCGGTCAAATCACCCTACATCATCATGGAAGACATTG AAGGTGTCCCTCTCCTCCACGACTGGTACAAAATCCGCGGGGACCCCGTCCGACATGCAATGCGGCGATTCGTCA CTGATGTCAGGCAAATGTTATGCATCCCATTCGACCAGATCGGAGGATTGTACTTTAAAGAGGACCTCCCTCCTC ACCTGCAAGAGCGGCCCTTGTTTTCACCCGCACTCTATCAAACCGAGACGGCTCTAAAAATGTTTGAGCCCGCTA TTGATAGATTTCGTGTTGGGCCTATTCCGGATTACACCTGGTGGAGGACGATGCATGACGAAGTAGGCGCAGACC GTGGCCCTTGTGAGTGTTATTCGACGCTACCACTTTGTCTGGCTGATCAATACCGACATAGGGAAGGATATCGGG GATATGCTCATCGCCGCTGTCAACTTGGAAAAAAGCGCCATTGCCAAATACCGACGTAATCCCTCCATTCTGAAA AAGACAGAGACCACCTCACTTGAAGAGCTTGACGTTCTCGAAGGGCTCCTCGAGAAGGTCGTAACCCTGGCGCCT TCATTGGCTTTGGCCTTGAGCAACATCCCAGATTTCATCAACTTCAATGTAGCCGTCCATGCTGATTTAGGGCCA AACAACGCTATGGTTCCTGCCGACAACGGCACAAACACACTCAGCCGTTTCGACCAGTTCACATACATCGACTGG CAGGGGACGTCCATCCTCCCCTTCCCCTTCCACTGCCACATACCCGAGCTCGTGCAATACAACGGCGACATCATC AAAGTCCCAGACGAGTATATAAACCAAATCCCAATTCCAGAAGGACTCGATCAACTTCCAGAAGACCAGCGAGAG TATATCATGGCACACCACCGCCTCGCCACCAGAGAATTGGCCTTTAAAAATGCCATGCTGGACAATTTCCATCCA TGGGCCGCTGCGAACGCGCACTGCCTCTTCCCTTATATCACCCAACTCCCCGACCGAGTCCTACGAGGTGTAACA CACGGGCCTATCTTGCTTCGAGAATGCCTAATGCGCATATCCTCGGAATGGGACGAGGAGCAGTGTGATGGACCG TGCCCCATTTCTTTCACGGAGGACGAAATTGCCGCGCACGACCGTGCAGCGGTCTTCCGTCATCTTTACTTTGTC AATACCAAGACTGCGCAAAAGCGTATCGATATGGATGGGGATGGGTGGGTGCCGAACGAAGAGTACGAGGAGGCG CGGGCGCGGTTTGAGAAGTATCGCGAGTCGTGGGATCCTAAGGTCTGCGGAGGACCCTGGCCTTTCCAAGATGGT GGTTACTCGTACTTCCTAGCCTAATACCTTTGTCTATATCTAGTTGTGAAATTTACCTCCTCTCACTAC |
| Length | 2019 |