CopciAB_503051
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_503051 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 503051 |
| Uniprot id | Functional description | other FunK1 protein kinase | |
| Location | scaffold_3:3475256..3478443 | Strand | - |
| Gene length (nt) | 3188 | Transcript length (nt) | 2795 |
| CDS length (nt) | 2520 | Protein length (aa) | 839 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4924 | 24.7 | 1.918E-35 | 146 |
| Lentinula edodes B17 | Lened_B_1_1_9238 | 24.8 | 8.817E-33 | 137 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_503051.T0 |
| Description | other FunK1 protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 217 | 681 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR040976 | Fungal-type protein kinase |
| IPR011009 | Protein kinase-like domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | other FunK1 protein kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
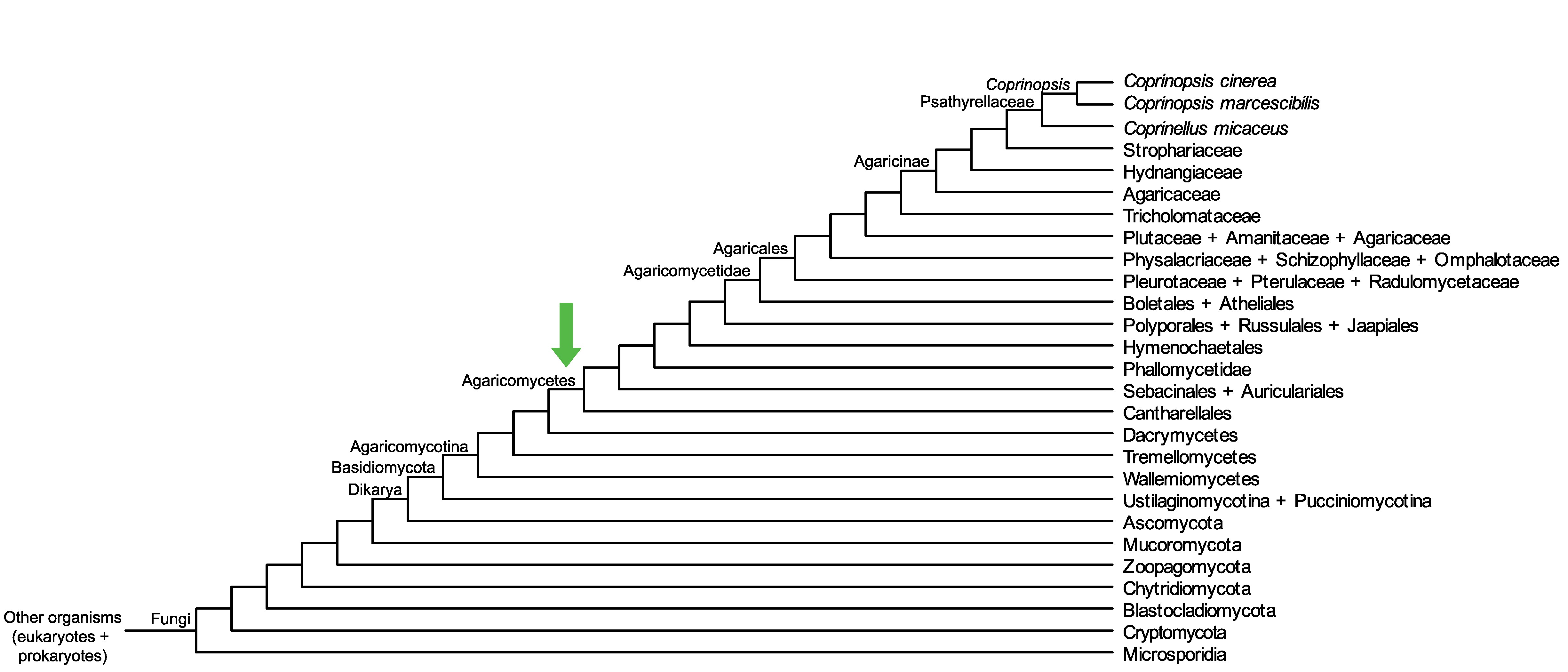
Conservation of CopciAB_503051 across fungi.
Arrow shows the origin of gene family containing CopciAB_503051.
Protein
| Sequence id | CopciAB_503051.T0 |
|---|---|
| Sequence |
>CopciAB_503051.T0 MERPRTRGAVRAEVRAQGSSHPQTPASSPIAHSAPVNDASVKIFTPRQDPDAVGDEKQTEIRDRIGRSMLNELVT CPCDAFFKEYLPPLPPGYPALTAQVSRMLTEKKIMTTGLFTAFRNITPSGLKARYERDKAHAKPGQVVPVVNEKT IFSSFGKIVTEIRNCLLCIPGLYVNPFPLALCPDTWLGSAVSGANFRIDCCTTSQVDSDGKPDLEGKRLHVRNIV TPMEWKISSSERRSNRLQVTAAANHIMNDDVRRMFLYGISVENERVTLWYYSRSHSVMSKSFDFVHEPHRLVELF IRLFCAKKEQLGFDPCISIPKAANGKQFLYKLPPTPEAKGDKPKGKRRYFLTTEIVDEYRSTRIAGRSTRVWKVV EVKGSSLELVDPNAEPLILKDAWITHDADTEKEIQDKVFGDIEKFASRDTVWASDELLKKFPKAALKSFEKALEN YTNYFSLIRHHYVGKPNKELSKEACWTALDVFFQVKPERPQTTTSTQRSGDKSGSNKRSVTKDGDILKQREYRAF ESKRRCFFVYELVCTRVSNLPTLGDAMDVLYQAYYALMMMFCAGWIHRDISHGNILAYKTPDGQWQVKLSDLEYA RKFPPEDGKVRTDPKTGTPYFMAHELLASAVLYEPKTTRPTKYFAPGEILPEREPKQGLKRAVMHNLQHDLESIW WMALWLSTMRTDLQDAYYAAKPIFQGSITLSTERYNCFTLGFGDEFQAKMDPSLQNIITELDELRDLLLSEYRGR HPDDACDLRKYSDACAGFLNFFAFINPSVGTWGPLALKDPSWLADPKPEEEEPSRIQPEPADRSTAVAELEASRS RTTYSIQEEAEPVL |
| Length | 839 |
Coding
| Sequence id | CopciAB_503051.T0 |
|---|---|
| Sequence |
>CopciAB_503051.T0 ATGGAAAGACCGAGAACCCGTGGAGCTGTCCGAGCTGAGGTGCGGGCACAGGGAAGCAGTCACCCACAGACTCCT GCCTCCTCGCCTATTGCTCACAGCGCCCCTGTTAACGATGCCAGTGTGAAGATATTCACCCCGCGTCAGGACCCC GATGCCGTAGGGGACGAAAAGCAAACCGAGATTCGCGATAGAATTGGGAGGAGTATGCTAAATGAACTCGTCACC TGCCCTTGCGATGCTTTCTTCAAAGAATACCTTCCTCCCCTTCCACCAGGATATCCGGCCTTGACCGCTCAAGTG TCTCGGATGTTAACCGAAAAAAAAATCATGACCACCGGCCTTTTCACTGCGTTTCGGAATATCACCCCAAGTGGC CTCAAAGCCCGATACGAACGGGACAAGGCCCATGCCAAACCGGGCCAAGTTGTGCCAGTGGTCAACGAGAAGACA ATCTTCTCCTCGTTTGGAAAAATCGTCACCGAGATACGAAACTGTCTGCTCTGTATCCCTGGCCTTTATGTTAAC CCTTTCCCCCTTGCTCTGTGCCCTGACACGTGGCTGGGCTCCGCTGTCAGCGGCGCCAACTTCAGAATCGACTGT TGTACCACTTCACAAGTGGACAGCGACGGCAAGCCCGACCTCGAAGGCAAAAGGCTTCACGTCAGGAACATCGTA ACTCCAATGGAATGGAAAATCTCTTCTTCCGAAAGACGATCGAATCGATTGCAGGTCACAGCTGCGGCTAACCAT ATCATGAACGACGATGTCCGCCGAATGTTCCTCTATGGTATATCCGTTGAGAATGAGAGAGTGACACTCTGGTAT TACTCTCGCTCTCATTCCGTCATGTCGAAATCCTTCGACTTTGTTCACGAACCCCATCGATTGGTCGAACTATTT ATCCGACTATTTTGCGCCAAAAAGGAGCAACTGGGGTTCGATCCCTGCATCTCCATCCCCAAGGCCGCCAATGGC AAACAGTTTTTATACAAATTGCCACCTACCCCAGAAGCGAAAGGCGACAAACCGAAAGGCAAGCGCCGATATTTT TTGACAACAGAGATAGTCGACGAGTACCGTTCCACTCGCATCGCTGGACGCTCCACCCGCGTATGGAAGGTAGTG GAAGTCAAGGGGTCGAGCCTTGAGCTTGTAGATCCCAACGCCGAGCCACTAATCCTAAAGGACGCCTGGATCACT CACGATGCTGACACCGAGAAGGAGATTCAGGACAAGGTTTTCGGCGACATCGAAAAATTCGCTTCACGCGATACC GTATGGGCCTCCGACGAATTGTTGAAAAAGTTTCCCAAGGCTGCACTTAAATCGTTCGAGAAGGCACTCGAAAAC TACACCAACTACTTCTCGCTCATTCGACATCATTATGTGGGGAAACCAAACAAGGAGTTGTCAAAAGAGGCGTGT TGGACCGCTCTGGACGTCTTTTTCCAGGTGAAGCCTGAAAGACCGCAGACGACGACATCAACCCAACGATCGGGT GACAAGAGCGGTTCGAACAAGAGAAGCGTCACAAAAGATGGCGACATCCTGAAGCAAAGAGAATATCGGGCGTTC GAGAGCAAGCGACGCTGTTTCTTTGTATATGAACTCGTCTGCACGCGCGTAAGCAATCTCCCTACATTGGGAGAC GCGATGGACGTGCTTTACCAAGCCTATTATGCGTTGATGATGATGTTCTGCGCTGGGTGGATCCACAGAGATATT AGCCACGGCAATATACTGGCGTATAAGACCCCGGATGGTCAGTGGCAAGTCAAGCTCTCCGATCTGGAATATGCC AGGAAATTCCCTCCGGAGGATGGTAAGGTCCGCACTGACCCTAAAACGGGAACGCCATACTTTATGGCTCACGAG CTCCTGGCATCTGCGGTACTTTATGAGCCAAAGACTACCAGGCCCACGAAATATTTCGCCCCTGGCGAGATATTA CCCGAGCGCGAGCCGAAGCAGGGCCTTAAACGGGCCGTTATGCACAACTTGCAGCACGATCTGGAATCTATATGG TGGATGGCGCTGTGGCTCTCCACCATGCGAACGGACCTTCAAGACGCATACTATGCTGCTAAACCGATCTTCCAA GGCTCGATTACCCTCTCCACGGAACGCTACAACTGCTTCACGCTCGGATTCGGGGATGAGTTCCAGGCCAAGATG GATCCGTCTCTCCAAAACATCATCACTGAACTTGACGAGCTCCGAGATCTTCTGCTCTCGGAGTACAGGGGACGC CATCCAGACGACGCCTGCGACCTGCGGAAGTACAGCGATGCTTGCGCCGGCTTCCTCAATTTCTTTGCCTTCATT AATCCATCGGTGGGAACATGGGGCCCGCTTGCTCTCAAGGACCCATCATGGCTCGCCGACCCCAAGCCAGAGGAG GAGGAACCTTCTCGAATCCAACCCGAACCCGCCGACCGTTCCACTGCAGTTGCCGAGCTCGAAGCGTCGCGATCG AGGACGACCTATTCAATCCAAGAAGAAGCCGAACCGGTGCTTTGA |
| Length | 2520 |
Transcript
| Sequence id | CopciAB_503051.T0 |
|---|---|
| Sequence |
>CopciAB_503051.T0 ATCGACCACCTCCGCCCATTTCATAACCAGCGAATGGAAAGACCGAGAACCCGTGGAGCTGTCCGAGCTGAGGTG CGGGCACAGGGAAGCAGTCACCCACAGACTCCTGCCTCCTCGCCTATTGCTCACAGCGCCCCTGTTAACGATGCC AGTGTGAAGATATTCACCCCGCGTCAGGACCCCGATGCCGTAGGGGACGAAAAGCAAACCGAGATTCGCGATAGA ATTGGGAGGAGTATGCTAAATGAACTCGTCACCTGCCCTTGCGATGCTTTCTTCAAAGAATACCTTCCTCCCCTT CCACCAGGATATCCGGCCTTGACCGCTCAAGTGTCTCGGATGTTAACCGAAAAAAAAATCATGACCACCGGCCTT TTCACTGCGTTTCGGAATATCACCCCAAGTGGCCTCAAAGCCCGATACGAACGGGACAAGGCCCATGCCAAACCG GGCCAAGTTGTGCCAGTGGTCAACGAGAAGACAATCTTCTCCTCGTTTGGAAAAATCGTCACCGAGATACGAAAC TGTCTGCTCTGTATCCCTGGCCTTTATGTTAACCCTTTCCCCCTTGCTCTGTGCCCTGACACGTGGCTGGGCTCC GCTGTCAGCGGCGCCAACTTCAGAATCGACTGTTGTACCACTTCACAAGTGGACAGCGACGGCAAGCCCGACCTC GAAGGCAAAAGGCTTCACGTCAGGAACATCGTAACTCCAATGGAATGGAAAATCTCTTCTTCCGAAAGACGATCG AATCGATTGCAGGTCACAGCTGCGGCTAACCATATCATGAACGACGATGTCCGCCGAATGTTCCTCTATGGTATA TCCGTTGAGAATGAGAGAGTGACACTCTGGTATTACTCTCGCTCTCATTCCGTCATGTCGAAATCCTTCGACTTT GTTCACGAACCCCATCGATTGGTCGAACTATTTATCCGACTATTTTGCGCCAAAAAGGAGCAACTGGGGTTCGAT CCCTGCATCTCCATCCCCAAGGCCGCCAATGGCAAACAGTTTTTATACAAATTGCCACCTACCCCAGAAGCGAAA GGCGACAAACCGAAAGGCAAGCGCCGATATTTTTTGACAACAGAGATAGTCGACGAGTACCGTTCCACTCGCATC GCTGGACGCTCCACCCGCGTATGGAAGGTAGTGGAAGTCAAGGGGTCGAGCCTTGAGCTTGTAGATCCCAACGCC GAGCCACTAATCCTAAAGGACGCCTGGATCACTCACGATGCTGACACCGAGAAGGAGATTCAGGACAAGGTTTTC GGCGACATCGAAAAATTCGCTTCACGCGATACCGTATGGGCCTCCGACGAATTGTTGAAAAAGTTTCCCAAGGCT GCACTTAAATCGTTCGAGAAGGCACTCGAAAACTACACCAACTACTTCTCGCTCATTCGACATCATTATGTGGGG AAACCAAACAAGGAGTTGTCAAAAGAGGCGTGTTGGACCGCTCTGGACGTCTTTTTCCAGGTGAAGCCTGAAAGA CCGCAGACGACGACATCAACCCAACGATCGGGTGACAAGAGCGGTTCGAACAAGAGAAGCGTCACAAAAGATGGC GACATCCTGAAGCAAAGAGAATATCGGGCGTTCGAGAGCAAGCGACGCTGTTTCTTTGTATATGAACTCGTCTGC ACGCGCGTAAGCAATCTCCCTACATTGGGAGACGCGATGGACGTGCTTTACCAAGCCTATTATGCGTTGATGATG ATGTTCTGCGCTGGGTGGATCCACAGAGATATTAGCCACGGCAATATACTGGCGTATAAGACCCCGGATGGTCAG TGGCAAGTCAAGCTCTCCGATCTGGAATATGCCAGGAAATTCCCTCCGGAGGATGGTAAGGTCCGCACTGACCCT AAAACGGGAACGCCATACTTTATGGCTCACGAGCTCCTGGCATCTGCGGTACTTTATGAGCCAAAGACTACCAGG CCCACGAAATATTTCGCCCCTGGCGAGATATTACCCGAGCGCGAGCCGAAGCAGGGCCTTAAACGGGCCGTTATG CACAACTTGCAGCACGATCTGGAATCTATATGGTGGATGGCGCTGTGGCTCTCCACCATGCGAACGGACCTTCAA GACGCATACTATGCTGCTAAACCGATCTTCCAAGGCTCGATTACCCTCTCCACGGAACGCTACAACTGCTTCACG CTCGGATTCGGGGATGAGTTCCAGGCCAAGATGGATCCGTCTCTCCAAAACATCATCACTGAACTTGACGAGCTC CGAGATCTTCTGCTCTCGGAGTACAGGGGACGCCATCCAGACGACGCCTGCGACCTGCGGAAGTACAGCGATGCT TGCGCCGGCTTCCTCAATTTCTTTGCCTTCATTAATCCATCGGTGGGAACATGGGGCCCGCTTGCTCTCAAGGAC CCATCATGGCTCGCCGACCCCAAGCCAGAGGAGGAGGAACCTTCTCGAATCCAACCCGAACCCGCCGACCGTTCC ACTGCAGTTGCCGAGCTCGAAGCGTCGCGATCGAGGACGACCTATTCAATCCAAGAAGAAGCCGAACCGGTGCTT TGATTTTGGGGACGGGGTCTGGTGTTTTTCTAAAGCTTTCGATTGTCCTTTTTGCCTTTTTTTTTCGCTCTCGTT GTCCTGCTTTGTTCTCGCTATCCGTTTTTGCTGTCCCTCGCTTGCTTTTTCAACCTCGCTTTCTCGACGTGTGAC ACCATCACCTACCGTTGTAGTCGTAGAATCACTTTCGTTGTCAAAAACTTGTAGTAATGTGGACCGTATGTCAGT GAATGGCCTTTACCTCACTC |
| Length | 2795 |
Gene
| Sequence id | CopciAB_503051.T0 |
|---|---|
| Sequence |
>CopciAB_503051.T0 ATCGACCACCTCCGCCCATTTCATAACCAGCGAATGGAAAGACCGAGAACCCGTGGAGCTGTCCGAGCTGAGGTG CGGGCACAGGGAAGCAGTCACCCACAGTGGGTTGAATAATTTATTATTTGCCCAATTTCCTTCAGGGGCTCACGA TGGATTGGTTCAGGACTCCTGCCTCCTCGCCTATTGCTCACAGCGCCCCTGTTAACGATGCCAGTGTGAAGATAT TGTGCGTCCCACTTTGCATTTGAAAGACTCTCGATGAGCGCTGATGACTGGCAGCACCCCGCGTCAGGACCCCGA TGCCGTAGGGGACGAAAAGCAAACCGAGATTCGCGATAGAATTGGGAGGAGTATGCTAAATGAACTCGTCACCTG CCCTTGCGATGCTTTCTTCAAAGAATACCTTCCTCCCCTTCCACCAGGATATCCGGCCTTGACCGCTCAAGTGTC TCGGATGTTAACCGAAAAAAAAATCATGACCACCGGCCTTTTCACTGCGTTTCGGAATATCACCCCAAGTGGCCT CAAAGCCCGATACGAACGGGACAAGGCCCATGCCAAACCGGGCCAAGTTGTGCCAGTGGTCAACGAGAAGACAAT CTTCTCCTCGTTTGGAAAAATCGTCACCGAGATACGAAACTGTCTGCTCTGTATCCCTGGCCTTTATGTTAACCC TTTCCCCCTTGCTCTGTGCCCTGACACGTGGCTGGGCTCCGCTGTCAGCGGCGCCAACTTCAGAATCGACTGTTG TACCACTTCACAAGTGGACAGCGACGGCAAGCCCGACCTCGAAGGCAAAAGGCTTCACGTCAGGAACATCGTAAC TCCAATGGAATGGAAAATCTCTTCTTCCGAAAGACGATCGGTCAGTACGATACCCACTTGCTGTCACAGTCTCAG TCTGTTCCTTACGACGGGTTGGTTACAGAATCGATTGCAGGTCACAGCTGCGGCTAACCATATCATGAACGACGA TGTCCGCCGAATGTTCCTCTATGGTGTATGTTTTCGCTCATATGGTCCAACGCGGCAGCTTCTGATCGGCGATTT GCAGATATCCGTTGAGAATGAGAGAGTGACACTCTGGTATTACTCTCGCTCTCATTCCGTCATGTCGAAATCCTT CGACTTTGTTCACGTGAGCCTGTTTCGCATCGGGAAGAGGCGGTCTATTCACATTCCACACAGGAACCCCATCGA TTGGTCGAACTATTTATCCGACTATTTTGCGCCAAAAAGGAGCAACTGGGGTTCGATCCCTGCATCTCCATCCCC AAGGCCGCCAATGGCAAACAGTTTTTATACAAATTGCCACCTACCCCAGAAGCGAAAGGCGACAAACCGAAAGGC AAGCGCCGATATTTTTTGACAACAGAGATAGTCGACGAGTACCGTTCCACTCGCATCGCTGGACGCTCCACCCGC GTATGGAAGGTAGTGGAAGTCAAGGGGTCGAGCCTTGAGCTTGTAGATCCCAACGCCGAGCCACTAATCCTAAAG GACGCCTGGATCACTCACGATGCTGACACCGAGAAGGAGATTCAGGACAAGGTTTTCGGCGACATCGAAAAATTC GCTTCACGCGATACCGTATGGGCCTCCGACGAATTGTTGAAAAAGTTTCCCAAGGCTGCACTTAAATCGTTCGAG AAGGCACTCGAAAACTACACCAACTACTTCTCGCTCATTCGACATCATTATGTGGGGAAACCAAACAAGGAGTTG TCAAAAGAGGCGTGTTGGACCGCTCTGGACGTCTTTTTCCAGGTGAAGCCTGAAAGACCGCAGACGACGACATCA ACCCAACGATCGGGTGACAAGAGCGGTTCGAACAAGAGAAGCGTCACAAAAGATGGCGACATCCTGAAGCAAAGA GAATATCGGGCGTTCGAGAGCAAGCGACGCTGTTTCTTTGTATATGAACTCGTCTGCACGCGCGTAAGCAATCTC CCTACATTGGGAGACGCGATGGACGTGCTTTACCAAGCCTATTATGGTGCGTCTGTCGCCTGTGGTTGTCCTTAG TCTTGGTCTAACCTCAGTCAAGCGTTGATGATGATGTTCTGCGCTGGGTGGATCCACAGAGATATTAGCCACGGC AATATACTGGCGTATAAGACCCCGGATGGTCAGTGGCAAGTCAAGCTCTCCGATCTGGAATATGCCAGGAAATTC CCTCCGGAGGATGGTAAGGTCCGCACTGACCCTAAAACGGTGCGTCGCTCTTGGGCTTGCAGTTATGTTGCAAAA CGTGACAGTCGTTTTCACTTCTAGGGAACGCCATACTTTATGGCTCACGAGCTCCTGGCATCTGCGGTACTTTAT GAGCCAAAGACTACCAGGCCCACGAAATATTTCGCCCCTGGCGAGATATTACCCGAGCGCGAGCCGAAGCAGGGC CTTAAACGGGCCGTTATGCACAACTTGCAGCACGATCTGGAATCTATATGGTGGATGGCGCTGTGGCTCTCCACC ATGCGAACGGACCTTCAAGACGCATACTATGCTGCTAAACCGATCTTCCAAGGCTCGATTACCCTCTCCACGGAA CGCTACAACTGCTTCACGCTCGGATTCGGGGATGAGTTCCAGGCCAAGATGGATCCGTCTCTCCAAAACATCATC ACTGAACTTGACGAGCTCCGAGATCTTCTGCTCTCGGAGTACAGGGGACGCCATCCAGACGACGCCTGCGACCTG CGGAAGTACAGCGATGCTTGCGCCGGCTTCCTCAATTTCTTTGCCTTCATTAATCCATCGGTGGGAACATGGGGC CCGCTTGCTCTCAAGGACCCATCATGGCTCGCCGACCCCAAGCCAGAGGAGGAGGAACCTTCTCGAATCCAACCC GAACCCGCCGACCGTTCCACTGCAGTTGCCGAGCTCGAAGCGTCGCGATCGAGGACGACCTATTCAATCCAAGAA GAAGCCGAACCGGTGCTTTGATTTTGGGGACGGGGTCTGGTGTTTTTCTAAAGCTTTCGATTGTCCTTTTTGCCT TTTTTTTTCGCTCTCGTTGTCCTGCTTTGTTCTCGCTATCCGTTTTTGCTGTCCCTCGCTTGCTTTTTCAACCTC GCTTTCTCGACGTGTGACACCATCACCTACCGTTGTAGTCGTAGAATCACTTTCGTTGTCAAAAACTTGTAGTAA TGTGGACCGTATGTCAGTGAATGGCCTTTACCTCACTC |
| Length | 3188 |