CopciAB_504019
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_504019 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | exgA | Synonyms | 504019 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_3:1094218..1095750 | Strand | + |
| Gene length (nt) | 1533 | Transcript length (nt) | 1170 |
| CDS length (nt) | 1170 | Protein length (aa) | 389 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Pleurotus eryngii ATCC 90797 | Pleery1_1445833 | 68.6 | 5.63E-184 | 572 |
| Agrocybe aegerita | Agrae_CAA7266842 | 64.5 | 1.296E-183 | 571 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB26208 | 64.6 | 1.596E-182 | 568 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1067593 | 66.2 | 6.848E-180 | 560 |
| Pleurotus ostreatus PC9 | PleosPC9_1_86868 | 65.9 | 7.329E-179 | 557 |
| Schizophyllum commune H4-8 | Schco3_2627090 | 65.5 | 1.549E-173 | 542 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_123281 | 62.3 | 9.616E-168 | 525 |
| Flammulina velutipes | Flave_chr09AA01159 | 61.2 | 1.057E-166 | 522 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_184098 | 58.6 | 2.053E-165 | 518 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_75231 | 58.8 | 2.139E-165 | 518 |
| Lentinula edodes NBRC 111202 | Lenedo1_1111530 | 59.3 | 5.14E-158 | 497 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_11327 | 59.3 | 5.101E-158 | 497 |
| Auricularia subglabra | Aurde3_1_1276417 | 58.1 | 7.552E-150 | 474 |
| Grifola frondosa | Grifr_OBZ72452 | 52 | 5.435E-138 | 439 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | exgA |
|---|---|
| Protein id | CopciAB_504019.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 75 | 312 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR017853 | Glycoside hydrolase superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01210 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_9 |
Transcription factor
| Group |
|---|
| No records |
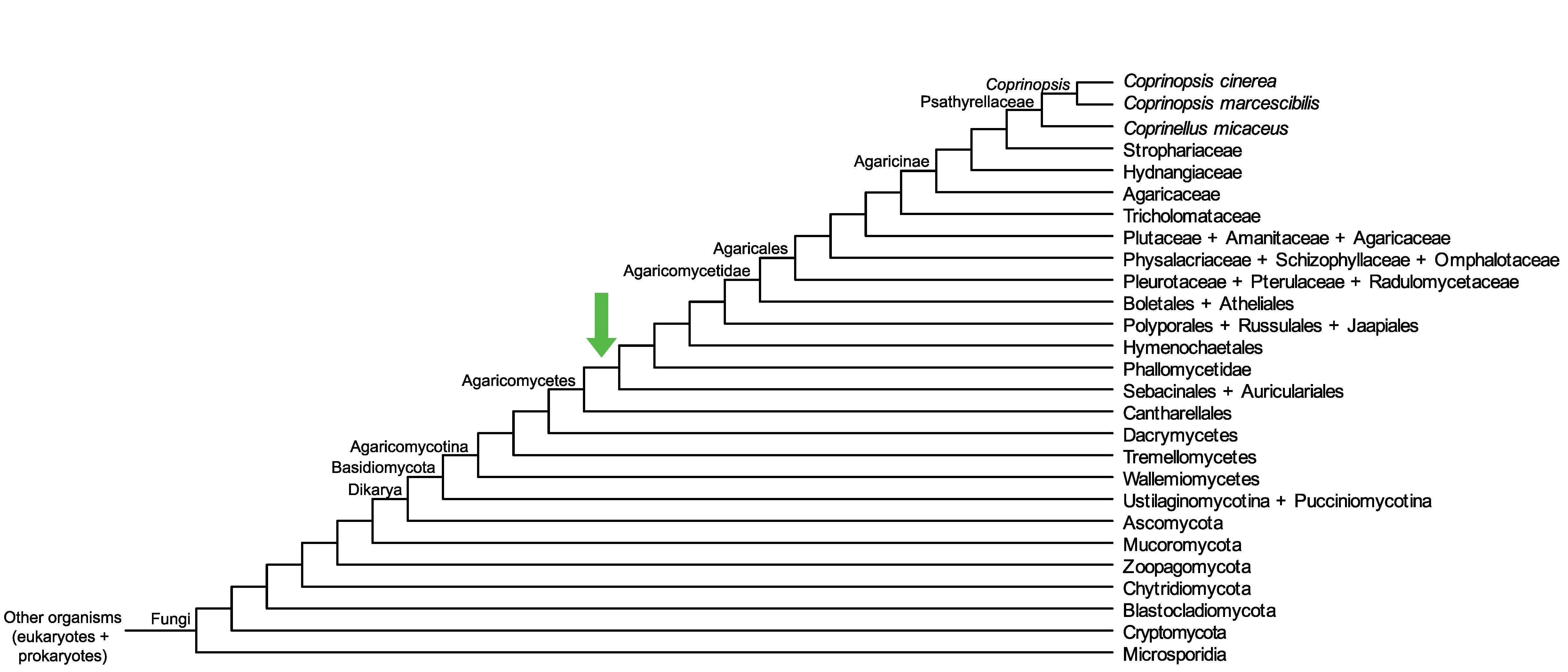
Conservation of CopciAB_504019 across fungi.
Arrow shows the origin of gene family containing CopciAB_504019.
Protein
| Sequence id | CopciAB_504019.T0 |
|---|---|
| Sequence |
>CopciAB_504019.T0 MEVLPAPSSTDAPEVQLPAFEYGRDKIRAVNLGGWLVLEPWITPSFFERTNNTDVIDEYTLGALVDRAAALEMLT QHWETWITEDDFIAIRAAGLTHVRIPLGFWSVPLTQDDVRTSVSSDPYIPGAWPYFLRGLTWARKHGVRVIVDLH GAPGSQNGYDNSGQRTSGPQWALQPHFVTHTVDVVRFIAANVGGLIDVLELLNEPAGFRGDDWAAVIREFWIEGY DAVRDAAGEDIHVMIGDAFLGVESWTDFLTPPRGHGVLMDFHEYQIFSHGELERSPQEHIDFACGYIDRLSSFAS SNLWTVVGEWSNAITDCARWLNGRGVGARWDGTWYDTDQYHDTCEGYTGSYHNFSEEYMAFLRRYWEVQVEIGES VQGWVFWTWKVCLS |
| Length | 389 |
Coding
| Sequence id | CopciAB_504019.T0 |
|---|---|
| Sequence |
>CopciAB_504019.T0 ATGGAAGTGCTCCCAGCGCCTTCAAGCACCGATGCACCAGAGGTCCAATTGCCCGCTTTCGAATACGGAAGAGAT AAAATCCGAGCAGTCAATCTGGGAGGATGGCTCGTTCTAGAGCCTTGGATAACACCCAGCTTCTTTGAAAGGACG AACAATACGGACGTCATAGACGAGTATACACTTGGGGCTCTTGTTGACCGTGCTGCTGCATTGGAAATGCTGACA CAACATTGGGAAACGTGGATAACGGAAGACGACTTTATCGCCATTCGTGCGGCAGGCCTTACCCATGTTCGGATA CCCCTCGGTTTCTGGTCCGTTCCCCTCACGCAAGACGACGTTCGCACTTCAGTTTCGTCCGACCCATACATTCCA GGCGCGTGGCCGTACTTCCTTCGTGGCCTCACCTGGGCACGCAAACACGGCGTGCGAGTCATTGTCGATCTGCAC GGTGCTCCAGGCTCACAGAACGGCTATGACAACTCCGGACAACGGACCTCCGGTCCTCAATGGGCTCTCCAACCG CACTTTGTGACGCATACCGTCGACGTTGTGCGCTTCATTGCGGCGAATGTTGGCGGGTTAATCGATGTACTAGAA CTCCTAAACGAACCCGCGGGCTTTAGAGGAGATGATTGGGCCGCAGTGATACGAGAGTTTTGGATAGAAGGGTAT GATGCTGTTAGGGATGCGGCTGGGGAGGACATTCATGTTATGATTGGGGATGCGTTTCTGGGAGTTGAGTCATGG ACTGATTTCCTGACACCTCCAAGAGGACATGGAGTTTTGATGGACTTCCACGAATACCAAATATTCAGTCACGGA GAGCTGGAACGATCACCACAAGAACATATCGACTTTGCGTGCGGATATATCGACCGATTGTCTAGCTTTGCGTCG TCGAACCTATGGACAGTCGTTGGTGAATGGTCGAACGCGATAACCGATTGCGCTCGCTGGTTAAATGGAAGAGGA GTTGGTGCTCGGTGGGATGGCACCTGGTACGACACAGACCAATATCACGACACCTGCGAGGGGTACACCGGGAGC TACCATAACTTCTCGGAAGAGTACATGGCATTTCTTAGAAGGTATTGGGAGGTTCAAGTCGAGATCGGAGAAAGC GTCCAAGGCTGGGTGTTTTGGACATGGAAGGTGTGTCTATCTTAA |
| Length | 1170 |
Transcript
| Sequence id | CopciAB_504019.T0 |
|---|---|
| Sequence |
>CopciAB_504019.T0 ATGGAAGTGCTCCCAGCGCCTTCAAGCACCGATGCACCAGAGGTCCAATTGCCCGCTTTCGAATACGGAAGAGAT AAAATCCGAGCAGTCAATCTGGGAGGATGGCTCGTTCTAGAGCCTTGGATAACACCCAGCTTCTTTGAAAGGACG AACAATACGGACGTCATAGACGAGTATACACTTGGGGCTCTTGTTGACCGTGCTGCTGCATTGGAAATGCTGACA CAACATTGGGAAACGTGGATAACGGAAGACGACTTTATCGCCATTCGTGCGGCAGGCCTTACCCATGTTCGGATA CCCCTCGGTTTCTGGTCCGTTCCCCTCACGCAAGACGACGTTCGCACTTCAGTTTCGTCCGACCCATACATTCCA GGCGCGTGGCCGTACTTCCTTCGTGGCCTCACCTGGGCACGCAAACACGGCGTGCGAGTCATTGTCGATCTGCAC GGTGCTCCAGGCTCACAGAACGGCTATGACAACTCCGGACAACGGACCTCCGGTCCTCAATGGGCTCTCCAACCG CACTTTGTGACGCATACCGTCGACGTTGTGCGCTTCATTGCGGCGAATGTTGGCGGGTTAATCGATGTACTAGAA CTCCTAAACGAACCCGCGGGCTTTAGAGGAGATGATTGGGCCGCAGTGATACGAGAGTTTTGGATAGAAGGGTAT GATGCTGTTAGGGATGCGGCTGGGGAGGACATTCATGTTATGATTGGGGATGCGTTTCTGGGAGTTGAGTCATGG ACTGATTTCCTGACACCTCCAAGAGGACATGGAGTTTTGATGGACTTCCACGAATACCAAATATTCAGTCACGGA GAGCTGGAACGATCACCACAAGAACATATCGACTTTGCGTGCGGATATATCGACCGATTGTCTAGCTTTGCGTCG TCGAACCTATGGACAGTCGTTGGTGAATGGTCGAACGCGATAACCGATTGCGCTCGCTGGTTAAATGGAAGAGGA GTTGGTGCTCGGTGGGATGGCACCTGGTACGACACAGACCAATATCACGACACCTGCGAGGGGTACACCGGGAGC TACCATAACTTCTCGGAAGAGTACATGGCATTTCTTAGAAGGTATTGGGAGGTTCAAGTCGAGATCGGAGAAAGC GTCCAAGGCTGGGTGTTTTGGACATGGAAGGTGTGTCTATCTTAA |
| Length | 1170 |
Gene
| Sequence id | CopciAB_504019.T0 |
|---|---|
| Sequence |
>CopciAB_504019.T0 ATGGAAGTGCTCCCAGCGCCTTCAAGCACCGATGCACCAGAGGTCCAATTGCCCGCTTTCGAATACGGAAGAGAT AAAATCCGAGCAGTCAATCTGTATGTCTATGTGGTTGGCTTTTTATGCAGTGATATTAACTTCAATCCAGGGGAG GATGGCTCGTTCTAGAGGTACTGAAACTCCGCGCCTTTCCCCTCCGGATCACAGCTAAACCACTCGTAGCCTTGG ATAACACCCAGCTTCTTTGAAAGGACGAACAATACGGACGTCATAGACGAGTATACACTTGGGGCTCTTGTTGAC CGTGCTGCTGCATTGGAAATGCTGACACAACATTGGGAAACGGTGTGTCTCCCTCTTTCTCTCTGTTTGTCCGCC TAAGTTCCGCTTTCTAGTGGATAACGGAAGACGACTTTATCGCCATTCGTGCGGCAGGCCTTACCCATGTTCGGT GAGTCGCCATCATTTCCCCTGTTACCCACCTTGCTCATGGAGTCACTCAGGATACCCCTCGGTTTCTGGTCCGTT CCCCTCACGCAAGACGACGTTCGCACTTCAGTTTCGTCCGACCCATACATTCCAGGCGCGTGGCCGTACTTCCTT CGTGGCCTCACCTGGGCACGCAAACACGGCGTGCGAGTCATTGTCGATCTGCACGGTGCTCCAGGCTCACAGAAC GGCTATGACAACTCCGGACAACGGACCTCCGGTCCTCAATGGGCTCTCCAACCGCACTTTGTGACGCATACCGTC GACGTTGTGCGCTTCATTGCGGCGAATGTTGGCGGGTTAATCGATGTACTAGAACTCCTAAACGAACCCGCGGGC TTTAGAGGAGATGATTGGGCCGCAGTGATACGAGAGTTTTGGATAGAAGGGTATGATGCTGTTAGGGATGCGGCT GGGGAGGACATTCATGTTATGATTGGGGATGCGTTTCTGGGAGTTGAGGTACATCGTCTAGATCCCCTTCCCGGC TGAACAATCGCTGATGCCCTGTATCCATAGTCATGGACTGATTTCCTGACACCTCCAAGAGGACATGGAGTTTTG ATGGACTTCGTGCGTACATCGTTCTCTGTTCATTCCCTCCATGCTGACCCTTTCACAGCACGAATACCAAATATT CAGTCACGGAGAGCTGGAACGATCACCACAAGAACATATCGACTTTGCGTGCGGATATATCGACCGATTGTCTAG CTTTGCGTCGTCGAACCTATGGACAGTCGTTGGTGAATGGTCGAACGCGATAACCGATTGCGCTCGCTGGTTAAA TGGAAGAGGAGTTGGTGCTCGGTGGGATGGCACCTGGTACGACACAGACCAATATCACGACACCTGCGAGGGGTA CACCGGGAGCTACCATAACTTCTCGGAAGAGTACATGGCATTTCTTAGAAGGTAAGTGTCGACATCCCTGTTGCT ATCTACGCCTGGTTACCACTCGTTCTTAGGTATTGGGAGGTTCAAGTCGAGATCGGAGAAAGCGTCCAAGGCTGG GTGTTTTGGACATGGAAGGTGTGTCTATCTTAA |
| Length | 1533 |