CopciAB_520581
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_520581 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | PKP2 | Synonyms | 520581 |
| Uniprot id | Functional description | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase | |
| Location | scaffold_5:1687233..1689346 | Strand | + |
| Gene length (nt) | 2114 | Transcript length (nt) | 1527 |
| CDS length (nt) | 1341 | Protein length (aa) | 446 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN9461_pkpB |
| Neurospora crassa | stk-24 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB22502 | 63.6 | 1.246E-177 | 557 |
| Agrocybe aegerita | Agrae_CAA7259766 | 63.5 | 4.492E-170 | 535 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_72910 | 63 | 7.348E-170 | 534 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_203285 | 63 | 1.563E-169 | 533 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1076172 | 62.3 | 1.972E-168 | 530 |
| Pleurotus ostreatus PC9 | PleosPC9_1_83342 | 62.3 | 4.28E-168 | 529 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1473475 | 61.8 | 2.528E-167 | 527 |
| Grifola frondosa | Grifr_OBZ70446 | 63.8 | 5.553E-166 | 523 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_115126 | 61 | 4.552E-159 | 503 |
| Auricularia subglabra | Aurde3_1_1272986 | 57.8 | 1.937E-157 | 499 |
| Schizophyllum commune H4-8 | Schco3_2621253 | 56.5 | 3.008E-157 | 498 |
| Flammulina velutipes | Flave_chr11AA00693 | 58.5 | 1.202E-156 | 496 |
| Lentinula edodes NBRC 111202 | Lenedo1_1201508 | 58.4 | 3.138E-155 | 492 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | PKP2 |
|---|---|
| Protein id | CopciAB_520581.T0 |
| Description | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF10436 | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase | IPR018955 | 48 | 209 |
| Pfam | PF02518 | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase | IPR003594 | 262 | 436 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR003594 | Histidine kinase/HSP90-like ATPase |
| IPR036890 | Histidine kinase/HSP90-like ATPase superfamily |
| IPR039028 | PDK/BCKDK protein kinase |
| IPR018955 | Branched-chain alpha-ketoacid dehydrogenase kinase/Pyruvate dehydrogenase kinase, N-terminal |
| IPR036784 | Alpha-ketoacid/pyruvate dehydrogenase kinase, N-terminal domain superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K16732 |
EggNOG
| COG category | Description |
|---|---|
| T | Mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
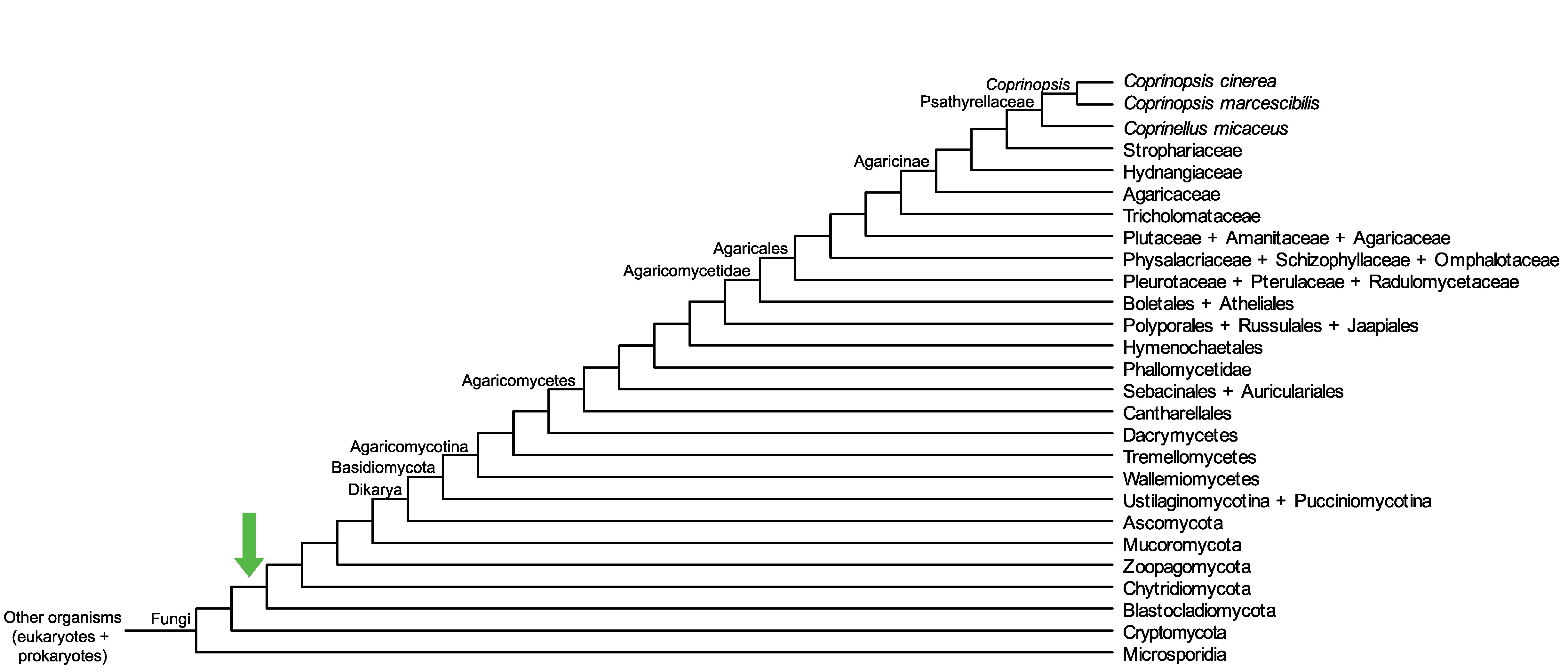
Conservation of CopciAB_520581 across fungi.
Arrow shows the origin of gene family containing CopciAB_520581.
Protein
| Sequence id | CopciAB_520581.T0 |
|---|---|
| Sequence |
>CopciAB_520581.T0 MTTFMVGRRVFNLTTTTPFLQSSLRYSSTFNFYQNRQLEQYANREAKPLTLRQLVFFGRHLTEEKILKSANYVRS ELPVRIAHRLRDLQALPYVVVTQEGVEKVYKLYWTAFEKFRTYPPITSIEENTKFCKFVGSLLDDHAVVIPNLSL GLSLSSPFLSPDKLDSFMRRMLVSRISRRVLAEHHIALSKTYLAKDSPAHEAEPRVGIIYTALDVKRCIDRCSTI LRDRPLWVHGNEDVRIDAWPEVEVEGHLDTKFAYIRDHLEYIVFELLKNAMSATVLKHHDSGSSLPPIRVTIVAG EDDISLRISDQGGGLTSVNAPTNDPMDLFSFSHIRNASRLEDSRLGALRTASEEGLRATVDEQLSRWQKHSYYQP KNRDKLEEHGTAPSQEIMNIVRSRIGIGLPMSNIYATYFGGSLELVSLDGWGTDVFLRLPKLGTNLEGIEV |
| Length | 446 |
Coding
| Sequence id | CopciAB_520581.T0 |
|---|---|
| Sequence |
>CopciAB_520581.T0 ATGACCACCTTCATGGTCGGACGACGCGTCTTCAATCTCACAACCACCACCCCATTTCTTCAATCGTCACTCAGA TATTCTTCAACGTTCAACTTTTATCAGAACAGGCAATTGGAGCAGTATGCTAATAGGGAGGCCAAACCTCTGACT TTGAGGCAACTTGTTTTCTTCGGGCGCCATCTTACGGAGGAGAAGATTCTTAAGAGTGCCAACTATGTTCGTTCT GAGTTGCCTGTCCGCATTGCCCATCGACTTCGGGACCTACAAGCGTTACCGTATGTTGTTGTAACACAGGAAGGG GTTGAGAAGGTTTATAAGCTGTACTGGACTGCATTTGAGAAGTTTCGGACATATCCACCAATAACATCAATAGAG GAGAACACCAAATTCTGCAAATTTGTAGGGTCGCTCCTGGATGACCATGCTGTTGTGATTCCAAACCTCTCTCTA GGCCTCTCCTTGTCGTCTCCATTCCTCTCGCCAGACAAGCTCGACAGCTTCATGCGCCGTATGCTCGTCTCCCGC ATATCCCGCCGTGTACTCGCAGAACACCATATCGCCCTGTCCAAGACGTACCTTGCTAAAGATTCTCCTGCTCAT GAAGCAGAACCTCGCGTTGGGATTATCTATACTGCTTTGGATGTTAAGCGGTGTATTGACAGATGCTCCACAATC TTGAGAGATAGACCGCTATGGGTACACGGGAACGAAGATGTCCGAATCGACGCCTGGCCGGAGGTCGAAGTCGAG GGACACCTAGATACCAAGTTTGCTTACATCCGGGATCACCTTGAATACATTGTATTTGAGCTCTTGAAGAATGCC ATGTCCGCCACTGTCCTCAAACATCACGACAGCGGCTCCTCGTTGCCACCCATCCGCGTTACAATCGTTGCTGGC GAGGACGATATCAGTCTGCGCATCTCGGACCAAGGCGGCGGTCTCACCTCCGTCAACGCCCCAACTAACGACCCC ATGGATCTGTTCTCTTTCTCCCATATACGGAACGCGTCGAGGTTGGAAGACTCCCGACTCGGTGCATTGCGCACT GCGAGCGAGGAAGGTCTGCGGGCGACGGTGGATGAACAACTCTCCCGTTGGCAGAAACATTCGTATTATCAACCG AAGAACCGGGATAAGTTGGAGGAGCATGGCACGGCGCCTTCTCAGGAAATCATGAATATTGTCCGTTCCAGGATT GGGATTGGCTTGCCTATGAGTAATATCTATGCGACATACTTTGGTGGCTCACTGGAGCTGGTATCGTTGGACGGT TGGGGTACCGATGTTTTTTTGCGACTTCCCAAGCTGGGGACCAATCTGGAGGGTATCGAGGTTTAG |
| Length | 1341 |
Transcript
| Sequence id | CopciAB_520581.T0 |
|---|---|
| Sequence |
>CopciAB_520581.T0 ACACAACTCCTCATACCGTCAGTCTTGAATGGGACAATCCTCGTAACGAGTAATGACATGACCACCTTCATGGTC GGACGACGCGTCTTCAATCTCACAACCACCACCCCATTTCTTCAATCGTCACTCAGATATTCTTCAACGTTCAAC TTTTATCAGAACAGGCAATTGGAGCAGTATGCTAATAGGGAGGCCAAACCTCTGACTTTGAGGCAACTTGTTTTC TTCGGGCGCCATCTTACGGAGGAGAAGATTCTTAAGAGTGCCAACTATGTTCGTTCTGAGTTGCCTGTCCGCATT GCCCATCGACTTCGGGACCTACAAGCGTTACCGTATGTTGTTGTAACACAGGAAGGGGTTGAGAAGGTTTATAAG CTGTACTGGACTGCATTTGAGAAGTTTCGGACATATCCACCAATAACATCAATAGAGGAGAACACCAAATTCTGC AAATTTGTAGGGTCGCTCCTGGATGACCATGCTGTTGTGATTCCAAACCTCTCTCTAGGCCTCTCCTTGTCGTCT CCATTCCTCTCGCCAGACAAGCTCGACAGCTTCATGCGCCGTATGCTCGTCTCCCGCATATCCCGCCGTGTACTC GCAGAACACCATATCGCCCTGTCCAAGACGTACCTTGCTAAAGATTCTCCTGCTCATGAAGCAGAACCTCGCGTT GGGATTATCTATACTGCTTTGGATGTTAAGCGGTGTATTGACAGATGCTCCACAATCTTGAGAGATAGACCGCTA TGGGTACACGGGAACGAAGATGTCCGAATCGACGCCTGGCCGGAGGTCGAAGTCGAGGGACACCTAGATACCAAG TTTGCTTACATCCGGGATCACCTTGAATACATTGTATTTGAGCTCTTGAAGAATGCCATGTCCGCCACTGTCCTC AAACATCACGACAGCGGCTCCTCGTTGCCACCCATCCGCGTTACAATCGTTGCTGGCGAGGACGATATCAGTCTG CGCATCTCGGACCAAGGCGGCGGTCTCACCTCCGTCAACGCCCCAACTAACGACCCCATGGATCTGTTCTCTTTC TCCCATATACGGAACGCGTCGAGGTTGGAAGACTCCCGACTCGGTGCATTGCGCACTGCGAGCGAGGAAGGTCTG CGGGCGACGGTGGATGAACAACTCTCCCGTTGGCAGAAACATTCGTATTATCAACCGAAGAACCGGGATAAGTTG GAGGAGCATGGCACGGCGCCTTCTCAGGAAATCATGAATATTGTCCGTTCCAGGATTGGGATTGGCTTGCCTATG AGTAATATCTATGCGACATACTTTGGTGGCTCACTGGAGCTGGTATCGTTGGACGGTTGGGGTACCGATGTTTTT TTGCGACTTCCCAAGCTGGGGACCAATCTGGAGGGTATCGAGGTTTAGCACCTGTCGCTGTTTCGTTTGGCGGTC GAGGACATTGCTTTATAATAATGAGACTATCTATCTGAATTGCTAGCGAACCGCACCTAATGTTACTAGTAGCAC GATTAGACAGAAGTAACTTTAAAGGTC |
| Length | 1527 |
Gene
| Sequence id | CopciAB_520581.T0 |
|---|---|
| Sequence |
>CopciAB_520581.T0 ACACAACTCCTCATACCGTCAGTCTTGAATGGGACAATCCTCGTAACGAGTAATGACATGACCACCTTCATGGTC GGACGACGCGTCTTCAATCTCACAACCACCACCCCATTTCTTCAATCGTCACTCAGATATTCTTCAACGTTCAAC TTTTATCAGAACAGGCAATTGGAGCAGTATGCTAATAGGGAGGCCAAACCTCTGACTTTGAGGCAACTTGTGAAT ATCACTTCTTGGTTGGGTATTGTGGTGTGTTTGATTTAATGTCTGGTTTACAGGTTTTCTTCGGGCGCCATCTTA CGGAGGAGAAGATTCTTAAGGTGGGAGAAACCACGAGTCAGTTTGGGAAGTGTGTCCTGAATCTAGGAAAGTAGA GTGCCAACTATGTTCGTTCTGAGTTGCCTGTCCGCATTGCCCATCGACTTCGGGACCTACAAGCGTTACCGTATG TTGTTGTAACACAGGAAGGGGTTGAGAAGGTTTATAAGGTGTGTTAGGGCTCCTTTCTATTCAGAGATGCAGATT CTAATCATCCTGTAGCTGTACTGGACTGCATTTGAGAAGTAAGGTCACTTGAACCTTTCGACCCGCCAGTCGCTC AGTGCTCAATCAGGTTTCGGACATATCCACCAATAACATCAATAGAGGAGAACACCAAATTCTGCAAATTTGTAG GGTCGCTCCTGGATGACCAGTAAGTCGGCTATGCACATTGCAAGCGAAATACTTAGCATCCCATCCCAGTGCTGT TGTGATTCCAAACCTCTCTCTAGGCCTCTCCTTGTCGTCTCCATTCCTCTCGCCAGACAAGCTCGACAGCTTCAT GCGCCGTATGCTCGTCTCCCGCATATCCCGCCGTGTACTCGCAGAACACCATATCGCCCTGTCCAAGACGTACCT TGCTAAAGATTCTCCTGCTCATGAAGCAGAACCTCGCGTTGGGATTATCTATACTGCTTTGGATGTTAAGCGGTG TATTGACAGATGCTCCACAATCTTGAGAGATAGACCGCTATGGGTACACGGGAACGAAGATGTCCGAATCGACGC CTGGCCGGAGGTCGAAGTCGAGGGACACCTAGATACCAAGTTTGCTTACATCCGGGATCACCTTGAGTAAGTCTA CATGCTATTCCGCAAGGCGATGTTAACCCTTCCTTCCTCAGATACATTGTATTTGAGCTCTTGAAGAATGTAGGT GCCATTATGGAAACCTTTAATTGCATTTGCTGAATCGTCCAATAGGCCATGTCCGCCACTGTCCTCAAACATCAC GACAGCGGCTCCTCGTTGCCACCCATCCGCGTTACAATCGTTGCTGGCGAGGACGATATCAGTCTGCGCATCTCG GACCAAGGTATGTACAAGGACGGAGTGTGTTGTGACCGCTTTTGACGGATCCTTCTTCTCCCTCTATCATAGGCG GCGGTCTCACCTCCGTCAACGCCCCAACTAACGACCCCATGGATCTGTTCTCTTTCTCCCATATACGGAACGCGT CGAGGTTGGAAGACTCCCGACTCGGTGCATTGCGCACTGCGAGCGAGGAAGGTCTGCGGGCGACGGTGGATGAAC AACTCTCCCGTTGGCAGAAACATTCGTATTATCAACCGAAGAACCGGGATAAGTTGGAGGAGCATGGCACGGCGC CTTCTCAGGAAATCATGAATATTGTCCGTTCCAGGATTGGGATTGGCTTGCCTATGAGTAATATCTATGCGACGT ATGGCCTAGTCTCGTATCGTTATTGATATCGCCTGAATGATCCCTCTCTCTCTCCAGATACTTTGGTGGCTCACT GGAGCTGGTATCGTTGGACGGTTGGGGTACGTTTTTACTTATAACAGACCGGCGGTTACACTCACTCTTATGTAG GTACCGATGTTTTTTTGCGACTTCCCAAGCTGGTGAGTAAATTAGACGTCCGAGCTCCACGCAGCTAACTGTGCA ATCAGGGGACCAATCTGGAGGGTATCGAGGTTTAGCACCTGTCGCTGTTTCGTTTGGCGGTCGAGGACATTGCTT TATAATAATGAGACTATCTATCTGAATTGCTAGCGAACCGCACCTAATGTTACTAGTAGCACGATTAGACAGAAG TAACTTTAAAGGTC |
| Length | 2114 |