CopciAB_521771
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_521771 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 521771 |
| Uniprot id | Functional description | mitogen-activated protein kinase kinase kinase | |
| Location | scaffold_13:731369..733589 | Strand | + |
| Gene length (nt) | 2221 | Transcript length (nt) | 1789 |
| CDS length (nt) | 1524 | Protein length (aa) | 507 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264551 | 54.8 | 4.18E-173 | 547 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21804 | 54.7 | 5.554E-171 | 541 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1060192 | 48.6 | 1.175E-147 | 473 |
| Pleurotus ostreatus PC9 | PleosPC9_1_117216 | 48.6 | 5.627E-147 | 471 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1445648 | 48.2 | 7.313E-146 | 468 |
| Schizophyllum commune H4-8 | Schco3_2702315 | 45.7 | 6.19E-129 | 419 |
| Lentinula edodes B17 | Lened_B_1_1_1607 | 39 | 6.702E-123 | 401 |
| Flammulina velutipes | Flave_chr03AA00206 | 47.5 | 1.854E-121 | 397 |
| Lentinula edodes NBRC 111202 | Lenedo1_1172327 | 38.1 | 2.318E-119 | 391 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18436 | 35.9 | 1.899E-65 | 233 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_521771.T0 |
| Description | mitogen-activated protein kinase kinase kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 205 | 480 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| T | mitogen-activated protein kinase kinase kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
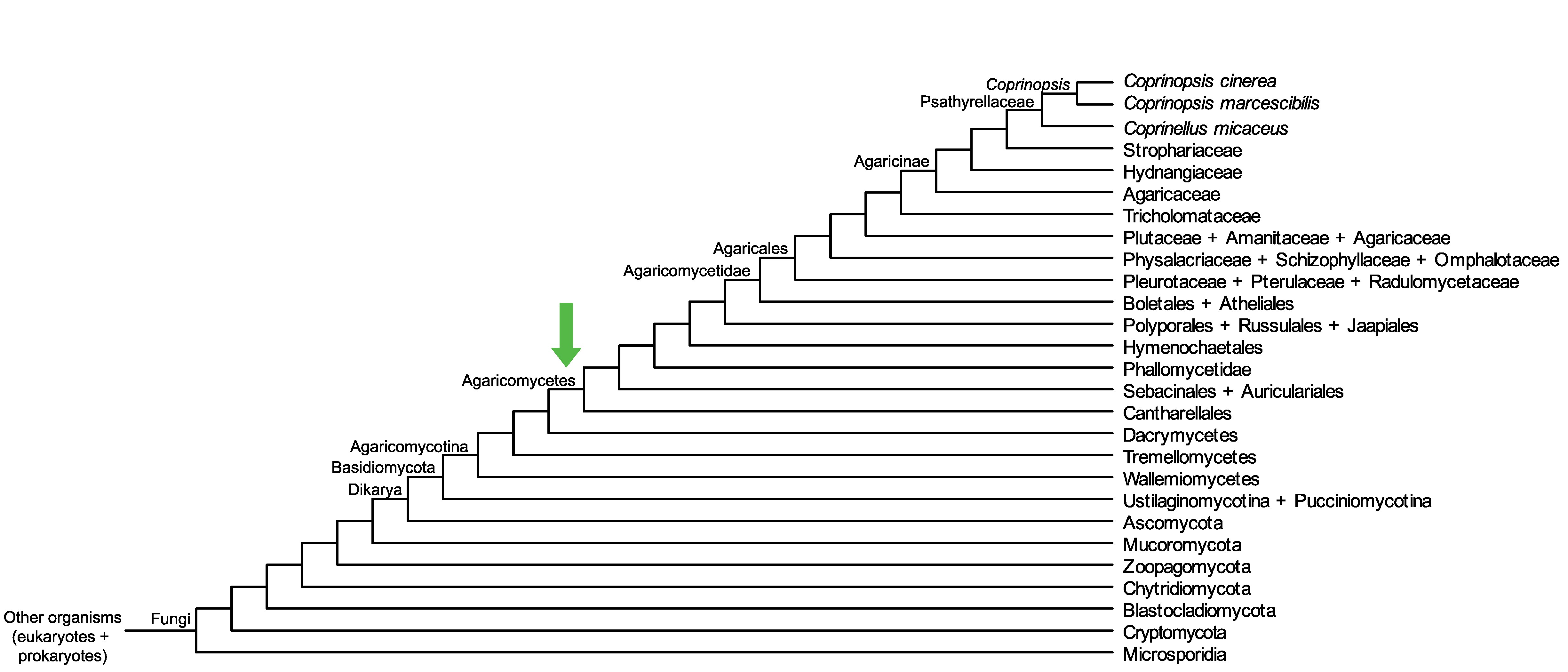
Conservation of CopciAB_521771 across fungi.
Arrow shows the origin of gene family containing CopciAB_521771.
Protein
| Sequence id | CopciAB_521771.T0 |
|---|---|
| Sequence |
>CopciAB_521771.T0 MDIYCWRLLEAEICVGVATDEQEYILPDCCYGALDEGFDEDREKCDFFFPISPAISVYLLGSSCFDDEPSFAPSE QPSSVSIHIGPESAIDVHLRNTMVLQTYPPYLIFSSLRGISLSVSSYYEFRWINEHQDYSRLRMRCRQKFMMEGV VKTLVIRESVESNGESQPPASPDSPVRMESPPGKDEVRVFDLTDEVQLKGNWAVAFGTFSDVWEGTWKDPIEKRD TAVAVKFLRQAMVKNVKERLIRRIQAEVATWHKLCHRNVSQFFGIVQSANSFGMVSPWYCNGIISEYVRKNPGVD RLKLLIQVASGIGHLHSFSPPIVHGDLKGGNILIDACGAAIITDFGLSKVMEETLTEVCEEDGGAVSGRGTSVFA GSTRWMAPELIMALVEDDDNGGPGPKITPMSDVYAFGSVCLEIATDELPYSHRRTDHAVTLDIIRGILPTRKDTC LVKGVDEDRFWQLLERCWSLDVTLRPTMIEALHWLDGLRNLNLLQNGTIHRRRQPLN |
| Length | 507 |
Coding
| Sequence id | CopciAB_521771.T0 |
|---|---|
| Sequence |
>CopciAB_521771.T0 ATGGATATCTATTGTTGGAGGTTGCTTGAAGCTGAGATTTGTGTTGGGGTCGCTACTGATGAGCAAGAGTACATC CTTCCGGATTGCTGCTACGGAGCTTTGGACGAGGGGTTTGACGAGGATAGGGAGAAATGTGACTTCTTCTTTCCT ATTAGCCCAGCCATTTCCGTCTACCTTCTGGGTAGCTCGTGCTTTGATGACGAACCTTCTTTCGCGCCGTCCGAA CAGCCGAGCTCCGTGTCCATCCATATTGGCCCAGAATCTGCCATCGACGTCCATCTGCGGAATACCATGGTCCTC CAGACGTACCCGCCGTACCTCATCTTCTCTTCCCTTCGTGGCATCTCCCTTTCGGTCTCCTCGTACTACGAATTC CGATGGATCAACGAGCATCAAGATTATTCGCGGTTGAGGATGCGGTGCAGGCAGAAGTTCATGATGGAGGGAGTG GTCAAGACTTTGGTCATCAGAGAAAGTGTAGAGTCGAACGGAGAGAGCCAGCCACCTGCCAGCCCGGACAGTCCT GTGCGGATGGAATCCCCGCCTGGGAAGGACGAGGTTCGGGTGTTCGACCTGACTGATGAAGTTCAGCTGAAAGGA AACTGGGCCGTCGCTTTTGGGACTTTCTCCGACGTTTGGGAGGGAACTTGGAAAGATCCTATTGAGAAGAGGGAC ACAGCGGTTGCTGTGAAGTTCCTGAGACAGGCGATGGTGAAGAATGTGAAGGAGCGACTCATTCGAAGAATACAA GCGGAGGTTGCAACGTGGCATAAACTGTGTCACCGCAACGTCAGCCAGTTCTTCGGTATCGTCCAATCGGCGAAT AGTTTTGGCATGGTCTCACCATGGTACTGCAATGGGATTATCTCCGAATACGTCAGGAAGAACCCTGGCGTAGAT CGACTGAAGCTGCTCATTCAAGTCGCTTCGGGAATAGGCCACCTTCATTCCTTCTCCCCTCCCATCGTTCATGGG GACCTTAAAGGAGGCAATATCCTCATTGATGCTTGCGGTGCAGCGATCATTACCGACTTCGGCTTGTCCAAGGTC ATGGAGGAAACCCTCACAGAGGTTTGTGAAGAGGATGGAGGGGCCGTGTCTGGCCGCGGTACATCGGTCTTTGCA GGTTCGACACGGTGGATGGCTCCCGAGCTAATAATGGCTCTTGTGGAAGATGATGACAATGGCGGGCCAGGTCCC AAGATCACTCCAATGAGTGATGTCTACGCGTTTGGAAGTGTCTGTCTGGAGATTGCGACGGACGAACTTCCCTAT TCCCATCGCAGAACAGACCATGCCGTGACGCTCGATATCATAAGAGGTATTTTACCCACTCGCAAGGATACTTGC CTAGTCAAAGGTGTGGACGAGGACAGATTCTGGCAGCTATTAGAACGGTGCTGGAGTCTGGATGTTACCCTACGG CCGACCATGATTGAGGCACTGCATTGGTTGGATGGACTCCGGAACTTGAACTTGTTACAGAACGGTACCATCCAC CGCCGGAGGCAACCCCTCAATTAA |
| Length | 1524 |
Transcript
| Sequence id | CopciAB_521771.T0 |
|---|---|
| Sequence |
>CopciAB_521771.T0 ACCCCGCACCACCAAGGTCCGGGAGACCCGACCATCTACTCTGAAGCCTTCCGGCCGTACCTCATCCGAACTAAA CGAAGGTGTGGACCTATCGCCCGACAACAAGTTTGTGGGATTGGTTCGAGAGGTTATGGATATCTATTGTTGGAG GTTGCTTGAAGCTGAGATTTGTGTTGGGGTCGCTACTGATGAGCAAGAGTACATCCTTCCGGATTGCTGCTACGG AGCTTTGGACGAGGGGTTTGACGAGGATAGGGAGAAATGTGACTTCTTCTTTCCTATTAGCCCAGCCATTTCCGT CTACCTTCTGGGTAGCTCGTGCTTTGATGACGAACCTTCTTTCGCGCCGTCCGAACAGCCGAGCTCCGTGTCCAT CCATATTGGCCCAGAATCTGCCATCGACGTCCATCTGCGGAATACCATGGTCCTCCAGACGTACCCGCCGTACCT CATCTTCTCTTCCCTTCGTGGCATCTCCCTTTCGGTCTCCTCGTACTACGAATTCCGATGGATCAACGAGCATCA AGATTATTCGCGGTTGAGGATGCGGTGCAGGCAGAAGTTCATGATGGAGGGAGTGGTCAAGACTTTGGTCATCAG AGAAAGTGTAGAGTCGAACGGAGAGAGCCAGCCACCTGCCAGCCCGGACAGTCCTGTGCGGATGGAATCCCCGCC TGGGAAGGACGAGGTTCGGGTGTTCGACCTGACTGATGAAGTTCAGCTGAAAGGAAACTGGGCCGTCGCTTTTGG GACTTTCTCCGACGTTTGGGAGGGAACTTGGAAAGATCCTATTGAGAAGAGGGACACAGCGGTTGCTGTGAAGTT CCTGAGACAGGCGATGGTGAAGAATGTGAAGGAGCGACTCATTCGAAGAATACAAGCGGAGGTTGCAACGTGGCA TAAACTGTGTCACCGCAACGTCAGCCAGTTCTTCGGTATCGTCCAATCGGCGAATAGTTTTGGCATGGTCTCACC ATGGTACTGCAATGGGATTATCTCCGAATACGTCAGGAAGAACCCTGGCGTAGATCGACTGAAGCTGCTCATTCA AGTCGCTTCGGGAATAGGCCACCTTCATTCCTTCTCCCCTCCCATCGTTCATGGGGACCTTAAAGGAGGCAATAT CCTCATTGATGCTTGCGGTGCAGCGATCATTACCGACTTCGGCTTGTCCAAGGTCATGGAGGAAACCCTCACAGA GGTTTGTGAAGAGGATGGAGGGGCCGTGTCTGGCCGCGGTACATCGGTCTTTGCAGGTTCGACACGGTGGATGGC TCCCGAGCTAATAATGGCTCTTGTGGAAGATGATGACAATGGCGGGCCAGGTCCCAAGATCACTCCAATGAGTGA TGTCTACGCGTTTGGAAGTGTCTGTCTGGAGATTGCGACGGACGAACTTCCCTATTCCCATCGCAGAACAGACCA TGCCGTGACGCTCGATATCATAAGAGGTATTTTACCCACTCGCAAGGATACTTGCCTAGTCAAAGGTGTGGACGA GGACAGATTCTGGCAGCTATTAGAACGGTGCTGGAGTCTGGATGTTACCCTACGGCCGACCATGATTGAGGCACT GCATTGGTTGGATGGACTCCGGAACTTGAACTTGTTACAGAACGGTACCATCCACCGCCGGAGGCAACCCCTCAA TTAAATCTCTGTTCTATTTATCCTGTCAAGCTATTCGTCATGCTATCTTGCTACTTTATACCCTGCTTTCGATAT CTGCTGACATCCTATCTGTCCTTTAGTTGGGCGAGTCAAATCCAAATTCAGGTTCTCGACACCC |
| Length | 1789 |
Gene
| Sequence id | CopciAB_521771.T0 |
|---|---|
| Sequence |
>CopciAB_521771.T0 ACCCCGCACCACCAAGGTCCGGGAGACCCGACCATCTACTCTGAAGCCTTCCGGCCGTACCTCATCCGAACTAAA CGAAGGTATCTCTTGAAGTGCTTTATCGACTTTTTGGAAGGAAATGACGCTCGTTCTCGCAATCACCTCCAAGGT GTGGACCTATCGCCCGACAACAAGTTTGTGGGATTGGTTCGAGAGGTTATGGATATCTATTGTTGGAGGTTGCTT GAAGCTGAGATTTGTGTTGGGGTCGCTACTGATGAGCAAGAGTACATCCTTCCGGATTGCTGCTACGGAGCTTTG GACGAGGGGTTTGACGAGGATAGGTGAGATCTGGTTGATTAAACTCTGGTATTGCTGTGGCTGACATCCATGCCA TTGGAATATAGGGAGAAATGTGACTTCTTCTTTCCTATTAGCCCAGCCATTTCCGTCTACCTTCTGGGTAGCTCG TGCTTTGATGACGAACCTTCTTTCGCGCCGTCCGAACAGCCGAGCTCCGTGTCCATCCATATTGGCCCAGAATCT GCCATCGACGTCCATCTGCGGAATACCATGGTCCTCCAGACGTACCCGCCGTACCTCATCTTCTCTTCCCTTCGT GGCATCTCCCTTTCGGTCTCCTCGTACTACGAATTCCGATGGATCAACGAGCATCAAGATTATTCGCGGTTGAGG ATGCGGTGCAGGCAGAAGTTCATGATGGAGGGAGTGGTCAAGACTTTGGTCATCAGAGAAAGTGTAGAGTCGAAC GGAGAGAGCCAGCCACCTGCCAGCCCGGACAGTCCTGTGCGGATGGAATCCCCGCCTGGGAAGGACGAGGTTCGG GTGTTCGACCTGACTGATGAAGTTCAGCTGAAAGGAAACTGGGCCGTCGCTTTTGGGACTTTCTCCGACGTTTGG GAGGGAACTTGGAAAGATCCTATTGAGAAGAGGGACACAGCGGTGAGTGGACCAGTCCCGATGGTGGATATAATA CCCATTGACTTCCATGCCAGGTTGCTGTGAAGTTCCTGAGACAGGCGATGGTGAAGAATGTGAAGGAGCGACTCA TTCGAGTGAGTAGGCACTAACAACAATTCGGGCCCCACTAACCAACCCTTCGTAGAGAATACAAGCGGAGGTTGC AACGTGGCATAAACTGTGTCACCGCAACGTCAGCCAGTTCTTCGGTATCGTCCAATCGGCGAATAGTTTTGGCAT GGTCTCACCATGGTACTGCAATGGGATTATCTCCGAATACGTCAGGAAGAACCCTGGCGTAGATCGACTGAAGCT GGTGCGCACTTCGAAGTCGATTAGTATCGCCATTTGGCCTGACCTTCTACCAGCTCATTCAAGTCGCTTCGGGAA TAGGCCACCTTCATTCCTTCTCCCCTCCCATCGTTCATGGGGACCTTAAAGGAGTTAGTTGACCTCTCCTCTAAT ACTGGCTGGACCTACGCGAGCGGCCCAAATCCCTTCTTGACCTGTGCTGATACCAAAGCCCACCATCTCCCATAG GGCAATATCCTCATTGATGCTTGCGGTGCAGCGATCATTACCGACTTCGGCTTGTCCAAGGTCATGGAGGAAACC CTCACAGAGGTTTGTGAAGAGGATGGAGGGGCCGTGTCTGGCCGCGGTACATCGGTCTTTGCAGGTTCGACACGG TGGATGGCTCCCGAGCTAATAATGGCTCTTGTGGAAGATGATGACAATGGCGGGCCAGGTCCCAAGATCACTCCA ATGAGTGATGTCTACGCGTTTGGAAGTGTCTGTCTGGAGGTGAGTGTTGATGAACAACCGTGCACTTGAAACTCA CGATCTCCTCCAGATTGCGACGGACGAACTTCCCTATTCCCATCGCAGAACAGACCATGCCGTGACGCTCGATAT CATAAGAGGTATTTTACCCACTCGCAAGGATACTTGCCTAGTCAAAGGTGTGGACGAGGACAGATTCTGGCAGCT ATTAGAACGGTGCTGGAGTCTGGATGTTACCCTACGGCCGACCATGATTGAGGCACTGCATTGGTTGGATGGACT CCGGAACTTGAACTTGTTACAGAACGGTACCATCCACCGCCGGAGGCAACCCCTCAATTAAATCTCTGTTCTATT TATCCTGTCAAGCTATTCGTCATGCTATCTTGCTACTTTATACCCTGCTTTCGATATCTGCTGACATCCTATCTG TCCTTTAGTTGGGCGAGTCAAATCCAAATTCAGGTTCTCGACACCC |
| Length | 2221 |