CopciAB_528616
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_528616 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 528616 |
| Uniprot id | Functional description | Centromere binding protein b | |
| Location | scaffold_7:969756..972416 | Strand | + |
| Gene length (nt) | 2661 | Transcript length (nt) | 2548 |
| CDS length (nt) | 1779 | Protein length (aa) | 592 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17905 | 36.5 | 2.028E-93 | 319 |
| Agrocybe aegerita | Agrae_CAA7261361 | 36.2 | 1.693E-63 | 230 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1106869 | 30.3 | 6.204E-59 | 216 |
| Flammulina velutipes | Flave_chr05AA00382 | 30.7 | 6.325E-56 | 207 |
| Grifola frondosa | Grifr_OBZ68949 | 32.7 | 5.785E-53 | 198 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1282558 | 35.8 | 4.647E-46 | 177 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_128049 | 29 | 1.773E-44 | 172 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_22879 | 28.3 | 6.801E-41 | 161 |
| Auricularia subglabra | Aurde3_1_1147515 | 36.1 | 1.021E-39 | 158 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_137362 | 38.4 | 2.042E-39 | 156 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_85261 | 38.9 | 9.294E-39 | 154 |
| Schizophyllum commune H4-8 | Schco3_2745584 | 27.9 | 1.885E-34 | 141 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87114 | 35.4 | 5.584E-28 | 120 |
| Lentinula edodes B17 | Lened_B_1_1_8771 | 30.3 | 1.452E-20 | 96 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_528616.T0 |
| Description | Centromere binding protein b |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF04218 | CENP-B N-terminal DNA-binding domain | IPR007889 | 265 | 306 |
| Pfam | PF03221 | Tc5 transposase DNA-binding domain | IPR006600 | 323 | 386 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR006600 | HTH CenpB-type DNA-binding domain |
| IPR009057 | Homeobox-like domain superfamily |
| IPR007889 | DNA binding HTH domain, Psq-type |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0003677 | DNA binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| B | Centromere binding protein b |
| D | Centromere binding protein b |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| helix-turn-helix |
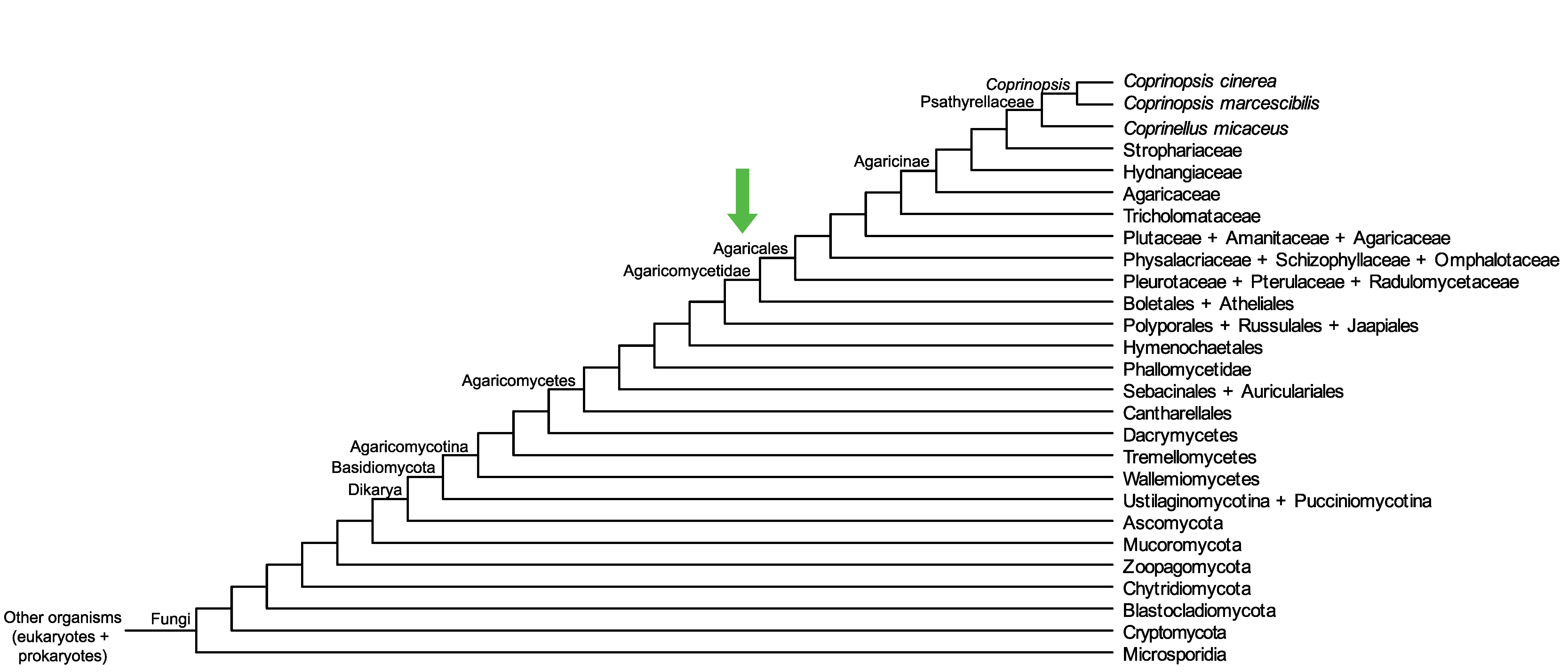
Conservation of CopciAB_528616 across fungi.
Arrow shows the origin of gene family containing CopciAB_528616.
Protein
| Sequence id | CopciAB_528616.T0 |
|---|---|
| Sequence |
>CopciAB_528616.T0 MDVHTDDNFACYRSDYRQPDRCVSTSGQTQSVASLPACDDTWIYPNYTVPQAFTGRNQMAPMYTTQRHPDQSYPS PSPSVPSPGPRVSPTLDLSSQTLGGLSGDNLSAEFCTRDSSLGPVRTSRSRQAATTYRLSRRTSISDPQLSLDTP ESRPSTPADDPSDLVPEQGLSLNINGSCPLQYQPATPTSLCSPYMTPYSEYPQNRPPSGHIDGRPPSPSLTSRSY MASGPSSPTAAYFPDVSPAATYHRNTRMRKHTKKKLDSLEKKRICLYHLDHQNARQEDIAEMFGIERSTVSKILK HKNQWLNVDDNAEAHAKNRPSKFPELEAVMRDFLQEALDKDIPLSDSLIRTRALEIAKGLGISEEKFKASSGWVE NFKHRHGIRSGKWTGGVKHVPAFDNPNPGLYGHTAMASRTYQPQLQMDSEDSTSGSPNRESSPELDDDTGDHGRT RPQPLHRPEDQDDSQWASSSSQRLSLSSAAVLPSPLSASSTNSADDGQSPQQCLPGGVDPNSSVVDMHASSTTYL PPESIPYAISTERRIPTLQEAEDAINILIAYIDSNPDRILEHSERTTLTTIKCALFQYASGVPFDRR |
| Length | 592 |
Coding
| Sequence id | CopciAB_528616.T0 |
|---|---|
| Sequence |
>CopciAB_528616.T0 ATGGACGTCCATACTGACGATAACTTTGCCTGCTACAGGTCGGACTACCGCCAACCAGACAGATGTGTTTCTACT AGTGGCCAGACCCAGTCCGTAGCCTCTTTGCCTGCCTGCGACGATACTTGGATATACCCAAACTATACCGTACCC CAGGCTTTCACTGGACGAAACCAAATGGCCCCAATGTACACGACTCAGCGTCATCCAGACCAGAGTTATCCCTCT CCATCGCCTTCAGTCCCTTCCCCGGGTCCTCGTGTATCTCCGACCCTCGATCTCTCATCCCAGACCCTGGGAGGC CTTTCAGGCGATAATTTGTCTGCCGAATTCTGCACCCGTGACTCCAGTCTTGGGCCAGTCAGGACTTCTCGCAGT CGACAAGCTGCTACGACCTACAGGCTTTCCCGCCGGACGTCGATCTCGGACCCGCAGCTCTCGCTGGATACCCCA GAGTCGCGTCCAAGCACCCCCGCCGACGACCCGTCGGACCTTGTTCCTGAACAGGGCCTATCATTGAACATCAAC GGCTCCTGCCCTCTTCAATACCAGCCAGCGACACCCACCTCTCTCTGCTCCCCATACATGACACCTTATAGCGAG TACCCGCAGAATCGCCCGCCATCGGGCCATATCGACGGTCGCCCGCCATCGCCTTCCCTCACTTCCAGGTCTTAC ATGGCCTCGGGTCCCAGCAGTCCAACAGCTGCCTACTTCCCCGATGTCTCCCCAGCCGCCACTTACCATCGAAAC ACTCGAATGAGGAAGCACACCAAGAAGAAGCTCGACTCGTTAGAGAAGAAGAGGATCTGTTTATATCACCTCGAC CACCAAAATGCACGTCAGGAAGACATCGCGGAAATGTTTGGCATCGAGCGCAGCACCGTCAGCAAGATCTTGAAA CACAAGAATCAGTGGCTCAATGTCGACGATAATGCAGAGGCCCATGCTAAGAATAGGCCATCCAAGTTCCCTGAG CTCGAGGCGGTGATGCGCGACTTCCTTCAAGAAGCTCTCGACAAAGACATCCCCCTCTCCGACTCCCTCATCCGC ACGCGTGCCTTGGAAATCGCAAAGGGGCTCGGCATCAGCGAAGAGAAGTTTAAGGCGAGCTCAGGGTGGGTCGAA AACTTCAAGCACCGACATGGCATCCGATCTGGCAAATGGACTGGCGGTGTCAAGCACGTTCCGGCGTTCGACAAT CCCAATCCAGGCCTCTATGGCCACACCGCCATGGCCTCTCGTACCTATCAGCCCCAGTTGCAGATGGACTCGGAG GATTCGACGTCTGGTAGCCCCAATCGAGAGTCTTCTCCCGAGCTCGACGATGATACCGGAGATCACGGGCGCACA AGACCCCAACCGCTACATCGTCCAGAAGACCAGGACGACTCCCAATGGGCCTCTTCGTCATCCCAACGCCTCTCC CTCTCCTCAGCAGCTGTACTCCCCTCGCCCCTTTCAGCATCGTCAACAAACTCTGCAGATGATGGGCAATCACCC CAGCAGTGCCTCCCCGGTGGAGTTGACCCTAATTCTTCGGTTGTCGACATGCATGCTTCATCCACCACCTACCTC CCACCGGAATCCATCCCCTACGCTATTTCGACTGAGCGCAGAATTCCAACTCTCCAGGAAGCGGAAGATGCGATC AACATTCTCATCGCCTACATCGACTCTAACCCCGACCGGATCCTCGAACACTCTGAACGAACGACATTAACGACG ATAAAATGCGCACTCTTCCAATACGCTAGTGGCGTCCCCTTTGATCGGCGCTGA |
| Length | 1779 |
Transcript
| Sequence id | CopciAB_528616.T0 |
|---|---|
| Sequence |
>CopciAB_528616.T0 AAATCCGCCATGGACGTCCATACTGACGATAACTTTGCCTGCTACAGGTCGGACTACCGCCAACCAGACAGATGT GTTTCTACTAGTGGCCAGACCCAGTCCGTAGCCTCTTTGCCTGCCTGCGACGATACTTGGATATACCCAAACTAT ACCGTACCCCAGGCTTTCACTGGACGAAACCAAATGGCCCCAATGTACACGACTCAGCGTCATCCAGACCAGAGT TATCCCTCTCCATCGCCTTCAGTCCCTTCCCCGGGTCCTCGTGTATCTCCGACCCTCGATCTCTCATCCCAGACC CTGGGAGGCCTTTCAGGCGATAATTTGTCTGCCGAATTCTGCACCCGTGACTCCAGTCTTGGGCCAGTCAGGACT TCTCGCAGTCGACAAGCTGCTACGACCTACAGGCTTTCCCGCCGGACGTCGATCTCGGACCCGCAGCTCTCGCTG GATACCCCAGAGTCGCGTCCAAGCACCCCCGCCGACGACCCGTCGGACCTTGTTCCTGAACAGGGCCTATCATTG AACATCAACGGCTCCTGCCCTCTTCAATACCAGCCAGCGACACCCACCTCTCTCTGCTCCCCATACATGACACCT TATAGCGAGTACCCGCAGAATCGCCCGCCATCGGGCCATATCGACGGTCGCCCGCCATCGCCTTCCCTCACTTCC AGGTCTTACATGGCCTCGGGTCCCAGCAGTCCAACAGCTGCCTACTTCCCCGATGTCTCCCCAGCCGCCACTTAC CATCGAAACACTCGAATGAGGAAGCACACCAAGAAGAAGCTCGACTCGTTAGAGAAGAAGAGGATCTGTTTATAT CACCTCGACCACCAAAATGCACGTCAGGAAGACATCGCGGAAATGTTTGGCATCGAGCGCAGCACCGTCAGCAAG ATCTTGAAACACAAGAATCAGTGGCTCAATGTCGACGATAATGCAGAGGCCCATGCTAAGAATAGGCCATCCAAG TTCCCTGAGCTCGAGGCGGTGATGCGCGACTTCCTTCAAGAAGCTCTCGACAAAGACATCCCCCTCTCCGACTCC CTCATCCGCACGCGTGCCTTGGAAATCGCAAAGGGGCTCGGCATCAGCGAAGAGAAGTTTAAGGCGAGCTCAGGG TGGGTCGAAAACTTCAAGCACCGACATGGCATCCGATCTGGCAAATGGACTGGCGGTGTCAAGCACGTTCCGGCG TTCGACAATCCCAATCCAGGCCTCTATGGCCACACCGCCATGGCCTCTCGTACCTATCAGCCCCAGTTGCAGATG GACTCGGAGGATTCGACGTCTGGTAGCCCCAATCGAGAGTCTTCTCCCGAGCTCGACGATGATACCGGAGATCAC GGGCGCACAAGACCCCAACCGCTACATCGTCCAGAAGACCAGGACGACTCCCAATGGGCCTCTTCGTCATCCCAA CGCCTCTCCCTCTCCTCAGCAGCTGTACTCCCCTCGCCCCTTTCAGCATCGTCAACAAACTCTGCAGATGATGGG CAATCACCCCAGCAGTGCCTCCCCGGTGGAGTTGACCCTAATTCTTCGGTTGTCGACATGCATGCTTCATCCACC ACCTACCTCCCACCGGAATCCATCCCCTACGCTATTTCGACTGAGCGCAGAATTCCAACTCTCCAGGAAGCGGAA GATGCGATCAACATTCTCATCGCCTACATCGACTCTAACCCCGACCGGATCCTCGAACACTCTGAACGAACGACA TTAACGACGATAAAATGCGCACTCTTCCAATACGCTAGTGGCGTCCCCTTTGATCGGCGCTGACCTCGGCTCGAC TTTCCCATTGTGCGTGCCTCTGTGGATCTGCTTCGACAGGAAGGCTCAGGCATGTCAGGTGCTTCTTACCTTCAC TAGGAGCAGAGCATCGCGGTAGCACCTCCAAAAGAGCGCGCTAGCTCTCGAGGTCGCTCAACCTCACAGGACCTT CGTTTTCATCTTCCATTCGTGCGGGTTTACGCTACATTCTCGTTTTCTCGATCACGACTAGCTCGTCACTCCCCC TGAAAGCATCATTTCGCTAGCAAGGTTTTGGTGTGCTGGACTACTTTTCGTACTCTGGTTTCTTTTTCTTTCTTC AATTATAGTTTCTTACTGGGGGTCCATCGAGCCGTAACAGCATTCGGCCTCGTCGTCATCTTTTTATCCTCGTGT ACATATATTTCATCATTGCCTCGTTTCCTGTCATACCCATGGGATTTGGATTTGGCTTTGGCTTCTTTTGCTCCA CTGCAGTCTCGACGGTCGCGCACCGCACTTCAATGCATTTCCACTAGTTTGTTACCCTTTTATGACTCTATGATT CCCCTCTGCCTCCATGAACCCCATGCGCTATGCTTTGTCCGTGTGGCCGGTATTAGCTCGGTACGAGTAGACACC CATCTTCAACACCATCCTATTTTCAGTAGTCACCACATCATCGAGCATTTTCTCCTTGTTTATCTACCTCTCGCC CATTGCCTCATTCAAGGACTATACCACCCTGTACAGTATATAATTATTACCGAATTACAGAGCCTCCGGAATC |
| Length | 2548 |
Gene
| Sequence id | CopciAB_528616.T0 |
|---|---|
| Sequence |
>CopciAB_528616.T0 AAATCCGCCATGGACGTCCATACTGACGATAACTTTGCCTGCTACAGGTCGGACTACCGCCAACCAGACAGATGT GTTTCTACTAGTGGCCAGACCCAGTCCGTAGCCTCTTTGCCTGCCTGCGACGATACTTGGATATACCCAAACTAT ACCGTACCCCAGGCTTTCACTGGACGAAACCAAATGGCCCCAATGTACACGACTCAGCGTCATCCAGACCAGAGT TATCCCTCTCCATCGCCTTCAGTCCCTTCCCCGGGTCCTCGTGTATCTCCGACCCTCGATCTCTCATCCCAGACC CTGGGAGGCCTTTCAGGCGATAATTTGTCTGCCGAATTCTGCACCCGTGACTCCAGTCTTGGGCCAGTCAGGACT TCTCGCAGTCGACAAGCTGCTACGACCTACAGGCTTTCCCGCCGGACGTCGATCTCGGACCCGCAGGTCCGTGGC CCGCCGTTTATACCTTTCGTTTTCCCCTGTCTAATCTACCTTTAGCTCTCGCTGGATACCCCAGAGTCGCGTCCA AGCACCCCCGCCGACGACCCGTCGGACCTTGTTCCTGAACAGGGCCTATCATTGAACATCAACGGCTCCTGCCCT CTTCAATACCAGCCAGCGACACCCACCTCTCTCTGCTCCCCATACATGACACCTTATAGCGAGTACCCGCAGAAT CGCCCGCCATCGGGCCATATCGACGGTCGCCCGCCATCGCCTTCCCTCACTTCCAGGTCTTACATGGCCTCGGGT CCCAGCAGTCCAACAGCTGCCTACTTCCCCGATGTCTCCCCAGCCGCCACTTACCATCGAAACACTCGAATGAGG AAGCACACCAAGAAGAAGCTCGACTCGTTAGAGAAGAAGAGGATCTGTTTATATCACCTCGACCACCAAAATGCA CGTCAGGAAGACATCGCGGAAATGTTTGGCATCGAGCGCAGCACCGTCAGCAAGATCTTGAAACACAAGAATCAG TGGCTCAATGTCGACGATAATGCAGAGGCCCATGCTAAGAATAGGTATGTCAAGTAGATCCATCGGGTTCCTTTC AAAGTCAAGCTAATTTTCCGGATCGCAGGCCATCCAAGTTCCCTGAGCTCGAGGCGGTGATGCGCGACTTCCTTC AAGAAGCTCTCGACAAAGACATCCCCCTCTCCGACTCCCTCATCCGCACGCGTGCCTTGGAAATCGCAAAGGGGC TCGGCATCAGCGAAGAGAAGTTTAAGGCGAGCTCAGGGTGGGTCGAAAACTTCAAGCACCGACATGGCATCCGAT CTGGCAAATGGACTGGCGGTGTCAAGCACGTTCCGGCGTTCGACAATCCCAATCCAGGCCTCTATGGCCACACCG CCATGGCCTCTCGTACCTATCAGCCCCAGTTGCAGATGGACTCGGAGGATTCGACGTCTGGTAGCCCCAATCGAG AGTCTTCTCCCGAGCTCGACGATGATACCGGAGATCACGGGCGCACAAGACCCCAACCGCTACATCGTCCAGAAG ACCAGGACGACTCCCAATGGGCCTCTTCGTCATCCCAACGCCTCTCCCTCTCCTCAGCAGCTGTACTCCCCTCGC CCCTTTCAGCATCGTCAACAAACTCTGCAGATGATGGGCAATCACCCCAGCAGTGCCTCCCCGGTGGAGTTGACC CTAATTCTTCGGTTGTCGACATGCATGCTTCATCCACCACCTACCTCCCACCGGAATCCATCCCCTACGCTATTT CGACTGAGCGCAGAATTCCAACTCTCCAGGAAGCGGAAGATGCGATCAACATTCTCATCGCCTACATCGACTCTA ACCCCGACCGGATCCTCGAACACTCTGAACGAACGACATTAACGACGATAAAATGCGCACTCTTCCAATACGCTA GTGGCGTCCCCTTTGATCGGCGCTGACCTCGGCTCGACTTTCCCATTGTGCGTGCCTCTGTGGATCTGCTTCGAC AGGAAGGCTCAGGCATGTCAGGTGCTTCTTACCTTCACTAGGAGCAGAGCATCGCGGTAGCACCTCCAAAAGAGC GCGCTAGCTCTCGAGGTCGCTCAACCTCACAGGACCTTCGTTTTCATCTTCCATTCGTGCGGGTTTACGCTACAT TCTCGTTTTCTCGATCACGACTAGCTCGTCACTCCCCCTGAAAGCATCATTTCGCTAGCAAGGTTTTGGTGTGCT GGACTACTTTTCGTACTCTGGTTTCTTTTTCTTTCTTCAATTATAGTTTCTTACTGGGGGTCCATCGAGCCGTAA CAGCATTCGGCCTCGTCGTCATCTTTTTATCCTCGTGTACATATATTTCATCATTGCCTCGTTTCCTGTCATACC CATGGGATTTGGATTTGGCTTTGGCTTCTTTTGCTCCACTGCAGTCTCGACGGTCGCGCACCGCACTTCAATGCA TTTCCACTAGTTTGTTACCCTTTTATGACTCTATGATTCCCCTCTGCCTCCATGAACCCCATGCGCTATGCTTTG TCCGTGTGGCCGGTATTAGCTCGGTACGAGTAGACACCCATCTTCAACACCATCCTATTTTCAGTAGTCACCACA TCATCGAGCATTTTCTCCTTGTTTATCTACCTCTCGCCCATTGCCTCATTCAAGGACTATACCACCCTGTACAGT ATATAATTATTACCGAATTACAGAGCCTCCGGAATC |
| Length | 2661 |