CopciAB_537608
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_537608 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 537608 |
| Uniprot id | Functional description | ||
| Location | scaffold_11:1653026..1655664 | Strand | - |
| Gene length (nt) | 2639 | Transcript length (nt) | 2466 |
| CDS length (nt) | 2133 | Protein length (aa) | 710 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14877 | 30.8 | 7.114E-58 | 216 |
| Agrocybe aegerita | Agrae_CAA7261224 | 29 | 1.651E-50 | 193 |
| Flammulina velutipes | Flave_chr08AA00914 | 31.1 | 1.836E-45 | 177 |
| Pleurotus ostreatus PC9 | PleosPC9_1_130681 | 30.2 | 1.668E-45 | 177 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1473715 | 26.2 | 1.789E-44 | 174 |
| Pleurotus ostreatus PC15 | PleosPC15_2_171810 | 26.4 | 1.521E-44 | 174 |
| Schizophyllum commune H4-8 | Schco3_2515142 | 26.5 | 7.915E-29 | 124 |
| Lentinula edodes B17 | Lened_B_1_1_16818 | 25 | 3.145E-22 | 102 |
| Lentinula edodes NBRC 111202 | Lenedo1_1072816 | 25 | 3.974E-22 | 102 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21072 | 28.6 | 7.983E-22 | 101 |
| Auricularia subglabra | Aurde3_1_1230869 | 28.3 | 0.0003 | 44 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_537608.T0 |
| Description |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 254 | 437 |
| Pfam | PF17667 | Fungal protein kinase | IPR040976 | 463 | 615 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR040976 | Fungal-type protein kinase |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| No records | |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
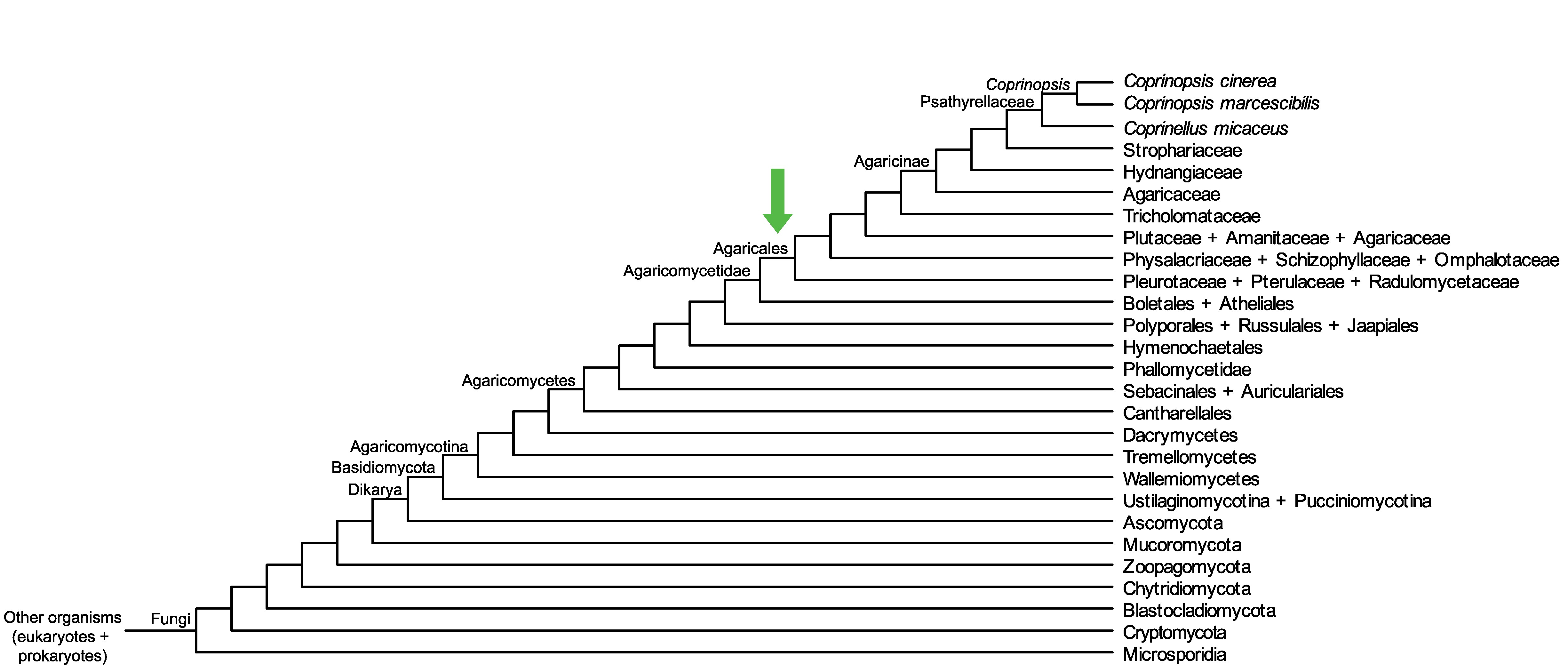
Conservation of CopciAB_537608 across fungi.
Arrow shows the origin of gene family containing CopciAB_537608.
Protein
| Sequence id | CopciAB_537608.T0 |
|---|---|
| Sequence |
>CopciAB_537608.T0 MLSPSNVRTSNGHDCGADSTRESIRRKLRGTYATDIPLIDFVKNVFDVEQSVISSLVGPGSTVTYSSEDVADSYR RLSSKGEAECCPYPRFFALVEDLLPAVFDVVKKERPYYGKEAYELGPEDLLDLTRGGRGVTYAKHGISRRKLDVL FLNGLDHASYLTEHPGEREQCEVPESRDSTSRPLSGDVRVSRAWTMGASGSSSIASSNFSSAKFTEFESGNDDAT PLTGTKRARSDSDATAQTSSAGGHVDKRPRTEGIDSERLAQLTECVIESLGSTERFYHVAFLVDNSNVCLAYVDH MVSLRSEWIDFIASPHLFSLMIVGFFFSTPRQAGCNPFLQATPVTEFDNSIVLPQTFPPVASPLRSFFAYKYPGS NDHEVAKPTSGGNKPLFYKPRGLFGRATTVYGTLATFVDRCVDNAALEGSWQPSIREESNIIDHLRKENSDFKVH FPEVLYSAAFSDRMGNIHGLDFEKRKNSRIIASPMYQTLWEVKTVEELQKAWLDAFKCHHVAWKTGEVLHRDVSE NNIMFLRRFFKNDETVVGILNDWDMASFLDEAYNLRQSTAKHRTGTLPFMSELLLFSDVEDSDDEEEEYQTVHHS YCHDVESFFWVLLWAFIHYPLDGTRRKTPDSLSQWISDDTEIVSFWKHTVLTKKKAVFNRMLRNHAFPHFQELKR DWLVPLYDLIAASRLTAPKVELTYEAVMKAIGEEA |
| Length | 710 |
Coding
| Sequence id | CopciAB_537608.T0 |
|---|---|
| Sequence |
>CopciAB_537608.T0 ATGTTATCTCCGTCTAACGTTCGAACCAGTAATGGTCACGACTGCGGAGCTGATTCCACCAGGGAGTCTATCCGA CGCAAACTTCGAGGGACCTATGCCACCGACATTCCCTTGATCGACTTCGTTAAGAATGTCTTTGATGTCGAGCAA TCAGTCATCTCTTCCCTCGTAGGCCCGGGATCAACGGTAACCTACTCGTCGGAGGATGTCGCCGATTCCTATCGC CGTCTTTCGAGCAAGGGAGAGGCTGAGTGTTGCCCATATCCCCGTTTCTTTGCATTAGTCGAAGACCTGTTGCCT GCCGTGTTTGACGTCGTCAAGAAGGAGCGTCCGTACTATGGGAAGGAGGCTTACGAGCTGGGCCCTGAGGATCTA CTTGACTTGACGCGGGGAGGTAGAGGGGTTACATACGCCAAACATGGGATCTCCAGACGGAAGCTCGACGTACTC TTCCTGAACGGACTTGATCATGCATCTTACCTCACCGAACACCCTGGAGAGCGCGAACAATGCGAGGTCCCAGAG TCCAGGGACAGTACGAGTCGTCCTCTCTCGGGTGACGTTCGAGTTTCGAGGGCCTGGACCATGGGCGCGTCCGGG TCGTCCTCCATCGCCTCATCCAACTTTTCGTCTGCAAAGTTTACTGAATTCGAATCTGGAAACGATGACGCGACA CCTTTGACTGGCACTAAACGCGCTCGTTCTGACTCTGACGCCACTGCTCAGACTAGTTCCGCGGGAGGACATGTG GACAAAAGGCCCAGAACGGAAGGGATCGACAGCGAGCGCCTAGCACAACTTACCGAATGCGTGATCGAGTCCTTG GGCAGCACTGAACGATTTTATCATGTCGCTTTCCTAGTCGACAATTCGAATGTCTGCCTCGCCTACGTTGATCAC ATGGTCTCTTTGCGCTCGGAGTGGATTGATTTCATTGCAAGTCCACATCTTTTCTCGTTGATGATCGTCGGTTTC TTCTTCTCCACTCCTCGACAAGCTGGCTGCAATCCGTTCCTTCAAGCGACCCCAGTTACTGAGTTCGACAACAGC ATTGTACTCCCACAGACCTTTCCGCCGGTAGCCTCCCCTCTCCGCTCTTTCTTCGCCTACAAATACCCTGGCTCC AACGACCACGAAGTCGCCAAACCAACCTCAGGCGGAAATAAGCCGTTGTTTTATAAACCTCGAGGTTTATTTGGA CGGGCGACCACCGTTTATGGGACTCTAGCAACCTTTGTCGACCGATGCGTTGACAATGCCGCCTTGGAAGGCAGT TGGCAACCCTCCATTCGGGAGGAGTCCAACATCATCGACCACCTCCGGAAGGAGAATTCTGACTTTAAGGTGCAC TTCCCAGAGGTGCTTTACAGCGCCGCCTTCAGTGACCGAATGGGTAACATCCATGGCCTTGACTTTGAGAAGCGG AAAAACTCGCGTATCATTGCATCCCCCATGTACCAGACACTGTGGGAAGTCAAGACTGTCGAGGAGTTACAGAAG GCCTGGCTTGATGCTTTCAAATGTCATCACGTCGCCTGGAAGACAGGGGAGGTCCTACATCGCGATGTCAGCGAG AACAACATCATGTTCTTGAGGAGGTTCTTCAAGAACGACGAAACTGTCGTCGGAATCCTCAATGACTGGGACATG GCTTCATTCCTGGACGAAGCGTACAATTTACGCCAGTCAACGGCCAAGCATCGGACGGGAACCCTTCCCTTCATG TCCGAACTTCTTCTATTTTCGGATGTGGAGGATTCAGATGATGAAGAGGAGGAGTACCAGACGGTTCATCACAGC TACTGTCACGACGTCGAGTCCTTCTTTTGGGTCCTTCTATGGGCTTTCATTCACTACCCACTCGATGGTACACGC AGGAAAACACCCGACTCTCTGAGCCAATGGATCTCCGACGACACCGAGATCGTCTCGTTCTGGAAACATACCGTT CTCACCAAGAAGAAGGCGGTCTTCAACAGGATGCTCAGAAACCACGCCTTTCCACACTTCCAGGAGCTCAAGCGC GATTGGCTTGTTCCTTTGTATGACCTCATTGCCGCGTCTCGGCTTACGGCTCCTAAGGTCGAGTTGACCTATGAA GCGGTGATGAAGGCTATTGGAGAGGAGGCGTAG |
| Length | 2133 |
Transcript
| Sequence id | CopciAB_537608.T0 |
|---|---|
| Sequence |
>CopciAB_537608.T0 CTTTTAACCACCATTGTTGCCAGTGTTTGGCTGGGTAATTCTCGTAATTATAAACCGCATCTCGGCTCGATGAAA CTCGTGTGCGTACATTGACTTTACTCTCCAAGAGCACTATGTTATCTCCGTCTAACGTTCGAACCAGTAATGGTC ACGACTGCGGAGCTGATTCCACCAGGGAGTCTATCCGACGCAAACTTCGAGGGACCTATGCCACCGACATTCCCT TGATCGACTTCGTTAAGAATGTCTTTGATGTCGAGCAATCAGTCATCTCTTCCCTCGTAGGCCCGGGATCAACGG TAACCTACTCGTCGGAGGATGTCGCCGATTCCTATCGCCGTCTTTCGAGCAAGGGAGAGGCTGAGTGTTGCCCAT ATCCCCGTTTCTTTGCATTAGTCGAAGACCTGTTGCCTGCCGTGTTTGACGTCGTCAAGAAGGAGCGTCCGTACT ATGGGAAGGAGGCTTACGAGCTGGGCCCTGAGGATCTACTTGACTTGACGCGGGGAGGTAGAGGGGTTACATACG CCAAACATGGGATCTCCAGACGGAAGCTCGACGTACTCTTCCTGAACGGACTTGATCATGCATCTTACCTCACCG AACACCCTGGAGAGCGCGAACAATGCGAGGTCCCAGAGTCCAGGGACAGTACGAGTCGTCCTCTCTCGGGTGACG TTCGAGTTTCGAGGGCCTGGACCATGGGCGCGTCCGGGTCGTCCTCCATCGCCTCATCCAACTTTTCGTCTGCAA AGTTTACTGAATTCGAATCTGGAAACGATGACGCGACACCTTTGACTGGCACTAAACGCGCTCGTTCTGACTCTG ACGCCACTGCTCAGACTAGTTCCGCGGGAGGACATGTGGACAAAAGGCCCAGAACGGAAGGGATCGACAGCGAGC GCCTAGCACAACTTACCGAATGCGTGATCGAGTCCTTGGGCAGCACTGAACGATTTTATCATGTCGCTTTCCTAG TCGACAATTCGAATGTCTGCCTCGCCTACGTTGATCACATGGTCTCTTTGCGCTCGGAGTGGATTGATTTCATTG CAAGTCCACATCTTTTCTCGTTGATGATCGTCGGTTTCTTCTTCTCCACTCCTCGACAAGCTGGCTGCAATCCGT TCCTTCAAGCGACCCCAGTTACTGAGTTCGACAACAGCATTGTACTCCCACAGACCTTTCCGCCGGTAGCCTCCC CTCTCCGCTCTTTCTTCGCCTACAAATACCCTGGCTCCAACGACCACGAAGTCGCCAAACCAACCTCAGGCGGAA ATAAGCCGTTGTTTTATAAACCTCGAGGTTTATTTGGACGGGCGACCACCGTTTATGGGACTCTAGCAACCTTTG TCGACCGATGCGTTGACAATGCCGCCTTGGAAGGCAGTTGGCAACCCTCCATTCGGGAGGAGTCCAACATCATCG ACCACCTCCGGAAGGAGAATTCTGACTTTAAGGTGCACTTCCCAGAGGTGCTTTACAGCGCCGCCTTCAGTGACC GAATGGGTAACATCCATGGCCTTGACTTTGAGAAGCGGAAAAACTCGCGTATCATTGCATCCCCCATGTACCAGA CACTGTGGGAAGTCAAGACTGTCGAGGAGTTACAGAAGGCCTGGCTTGATGCTTTCAAATGTCATCACGTCGCCT GGAAGACAGGGGAGGTCCTACATCGCGATGTCAGCGAGAACAACATCATGTTCTTGAGGAGGTTCTTCAAGAACG ACGAAACTGTCGTCGGAATCCTCAATGACTGGGACATGGCTTCATTCCTGGACGAAGCGTACAATTTACGCCAGT CAACGGCCAAGCATCGGACGGGAACCCTTCCCTTCATGTCCGAACTTCTTCTATTTTCGGATGTGGAGGATTCAG ATGATGAAGAGGAGGAGTACCAGACGGTTCATCACAGCTACTGTCACGACGTCGAGTCCTTCTTTTGGGTCCTTC TATGGGCTTTCATTCACTACCCACTCGATGGTACACGCAGGAAAACACCCGACTCTCTGAGCCAATGGATCTCCG ACGACACCGAGATCGTCTCGTTCTGGAAACATACCGTTCTCACCAAGAAGAAGGCGGTCTTCAACAGGATGCTCA GAAACCACGCCTTTCCACACTTCCAGGAGCTCAAGCGCGATTGGCTTGTTCCTTTGTATGACCTCATTGCCGCGT CTCGGCTTACGGCTCCTAAGGTCGAGTTGACCTATGAAGCGGTGATGAAGGCTATTGGAGAGGAGGCGTAGGGAT TTAGAACAGTTTTGGTGTCTTTTTTCTTTTCTTGTTTCAGTAATGCGCACTGGTCACCCAGATATCTTTAACCAC CTTCACATCTGTATTCTGACTGTAACTATTTATCACTGAGCTTCATCGAGATCTCTTATTTATTCAACAACCCCT TAATTATTACTACAATCCTGTACTCTCGTTACTGTACTTCACATTGAATTGATACATCATTCAGAC |
| Length | 2466 |
Gene
| Sequence id | CopciAB_537608.T0 |
|---|---|
| Sequence |
>CopciAB_537608.T0 CTTTTAACCACCATTGTTGCCAGTGTTTGGCTGGGTAATTCTCGTAATTATAAACCGCATCTCGGCTCGATGGTG ATTCCAACACTTCCTTCCCGACACACTCATCTCTAAGGACGTCCTCATCCAGAAACTCGTGTGCGTACATTGACT TTACTCTCCAAGAGGTATGTCGACTATGGTACGACCTCAGGTGCATGCTTGGGCTTAACTGTATTTTTACCTAGC ACTATGTTATCTCCGTCTAACGTTCGAACCAGTAATGGTCACGACTGCGGAGCTGATTCCACCAGGGAGTCTATC CGACGCAAACTTCGAGGGACCTATGCCACCGACATTCCCTTGATCGACTTCGTTAAGAATGTCTTTGATGTCGAG CAATCAGTCATCTCTTCCCTCGTAGGCCCGGGATCAACGGTAACCTACTCGTCGGAGGATGTCGCCGATTCCTAT CGCCGTCTTTCGAGCAAGGGAGAGGCTGAGTGTTGCCCATATCCCCGTTTCTTTGCATTAGTCGAAGACCTGTTG CCTGCCGTGTTTGACGTCGTCAAGAAGGAGCGTCCGTACTATGGGAAGGAGGCTTACGAGCTGGGCCCTGAGGAT CTACTTGACTTGACGCGGGGAGGTAGAGGGGTTACATACGCCAAACATGGGATCTCCAGACGGAAGCTCGACGTA CTCTTCCTGAACGGACTTGATCATGCATCTTACCTCACCGAACACCCTGGAGAGCGCGAACAATGCGAGGTCCCA GAGTCCAGGGACAGTACGAGTCGTCCTCTCTCGGGTGACGTTCGAGTTTCGAGGGCCTGGACCATGGGCGCGTCC GGGTCGTCCTCCATCGCCTCATCCAACTTTTCGTCTGCAAAGTTTACTGAATTCGAATCTGGAAACGATGACGCG ACACCTTTGACTGGCACTAAACGCGCTCGTTCTGACTCTGACGCCACTGCTCAGACTAGTTCCGCGGGAGGACAT GTGGACAAAAGGCCCAGAACGGAAGGGATCGACAGCGAGCGCCTAGCACAACTTACCGAATGCGTGATCGAGTCC TTGGGCAGCACTGAACGATTTTATCATGTCGCTTTCCTAGTCGACAATTCGAATGTCTGCCTCGCCTACGTTGAT CACATGGTCTCTTTGCGCTCGGAGTGGATTGATTTCATTGCAAGTCCACATCTTTTCTCGTTGATGATCGTCGGT TTCTTCTTCTCCACTCCTCGACAAGCTGGCTGCAATCCGTTCCTTCAAGCGACCCCAGTTACTGAGTTCGACAAC AGCATTGTACTCCCACAGACCTTTCCGCCGGTAGCCTCCCCTCTCCGCTCTTTCTTCGCCTACAAATACCCTGGC TCCAACGACCACGAAGTCGCCAAACCAACCTCAGGCGGAAATAAGCCGTTGTTTTATAAACCTCGAGGTTTATTT GGACGGGCGACCACCGTTTATGGGACTCTAGCAACCTTTGTCGACCGATGCGTTGACAATGCCGCCTTGGAAGGC AGTTGGCAACCCTCCATTCGGGAGGAGTCCAACATCATCGACCACCTCCGGAAGGAGAATTCTGACTTTAAGGTG CACTTCCCAGAGGTGCTTTACAGCGCCGCCTTCAGTGACCGAATGGGTAACATCCATGGCCTTGACTTTGAGAAG CGGAAAAACTCGCGTATCATTGCATCCCCCATGTACCAGACACTGTGGGAAGTCAAGACTGTCGAGGAGTTACAG AAGGCCTGGCTTGATGCTTTCAAATGTAAGTATTTTGTCACGCTCTCTCCTCGAATTGTTTCATACTGACGTATG TCCTGCAGGTCATCACGTCGCCTGGAAGACAGGGGAGGTCCTACATCGCGATGTCAGCGAGAACAACATCATGTT CTTGAGGAGGTTCTTCAAGAACGACGAAACTGTCGTCGGAATCCTCAATGACTGGGACATGGCTTCATTCCTGGA CGAAGCGTACAATTTACGCCAGTCAACGGCCAAGCATCGGACGGGAACCCTTCCCTTCATGTCCGAACTTCTTCT ATTTTCGGATGTGGAGGATTCAGATGATGAAGAGGAGGAGTACCAGACGGTTCATCACAGCTACTGTCACGACGT CGAGTCCTTCTTTTGGGTCCTTCTATGGGCTTTCATTCACTACCCACTCGATGGTACACGCAGGAAAACACCCGA CTCTCTGAGCCAATGGATCTCCGACGACACCGAGATCGTCTCGTTCTGGAAACATACCGTTCTCACCAAGAAGAA GGCGGTCTTCAACAGGATGCTCAGAAACCACGCCTTTCCACACTTCCAGGAGCTCAAGCGCGATTGGCTTGTTCC TTTGTATGACCTCATTGCCGCGTCTCGGCTTACGGCTCCTAAGGTCGAGTTGACCTATGAAGCGGTGATGAAGGC TATTGGAGAGGAGGCGTAGGGATTTAGAACAGTTTTGGTGTCTTTTTTCTTTTCTTGTTTCAGTAATGCGCACTG GTCACCCAGATATCTTTAACCACCTTCACATCTGTATTCTGACTGTAACTATTTATCACTGAGCTTCATCGAGAT CTCTTATTTATTCAACAACCCCTTAATTATTACTACAATCCTGTACTCTCGTTACTGTACTTCACATTGAATTGA TACATCATTCAGAC |
| Length | 2639 |