CopciAB_539376
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_539376 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | SCH9 | Synonyms | 539376 |
| Uniprot id | Functional description | Haspin like kinase domain | |
| Location | scaffold_5:2270032..2273888 | Strand | - |
| Gene length (nt) | 3857 | Transcript length (nt) | 3622 |
| CDS length (nt) | 2421 | Protein length (aa) | 806 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | stk-10 |
| Schizosaccharomyces pombe | sck1 |
| Saccharomyces cerevisiae | YHR205W_SCH9 |
| Candida albicans | SCH9 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7260185 | 65.3 | 2.21E-304 | 966 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB23235 | 64.9 | 1.221E-304 | 964 |
| Grifola frondosa | Grifr_OBZ70028 | 65.8 | 1.263E-291 | 926 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1063869 | 64.6 | 2.614E-291 | 925 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1385862 | 64.3 | 6.809E-292 | 924 |
| Lentinula edodes NBRC 111202 | Lenedo1_1035474 | 64.5 | 3.313E-297 | 922 |
| Pleurotus ostreatus PC9 | PleosPC9_1_76610 | 64.3 | 1.48E-291 | 917 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_115021 | 65.8 | 3.383E-286 | 890 |
| Schizophyllum commune H4-8 | Schco3_9874 | 62.8 | 4.183E-286 | 890 |
| Flammulina velutipes | Flave_chr11AA00390 | 60.9 | 8.172E-285 | 886 |
| Auricularia subglabra | Aurde3_1_110485 | 60.5 | 3.442E-283 | 882 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_135170 | 58.8 | 4.815E-279 | 869 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_113660 | 62.4 | 6.409E-256 | 802 |
| Lentinula edodes B17 | Lened_B_1_1_13175 | 60.6 | 6.07E-204 | 651 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_9646 | 60.6 | 7.612E-204 | 651 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | SCH9 |
|---|---|
| Protein id | CopciAB_539376.T0 |
| Description | Haspin like kinase domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd11651 | YPK1_N_like | - | 64 | 284 |
| Pfam | PF12330 | Haspin like kinase domain | - | 116 | 447 |
| Pfam | PF00168 | C2 domain | IPR000008 | 205 | 259 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 292 | 551 |
| Pfam | PF00433 | Protein kinase C terminal domain | IPR017892 | 572 | 699 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000719 | Protein kinase domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR017892 | Protein kinase, C-terminal |
| IPR000008 | C2 domain |
| IPR000961 | AGC-kinase, C-terminal |
| IPR035892 | C2 domain superfamily |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K08286 |
| K19800 |
EggNOG
| COG category | Description |
|---|---|
| T | Haspin like kinase domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
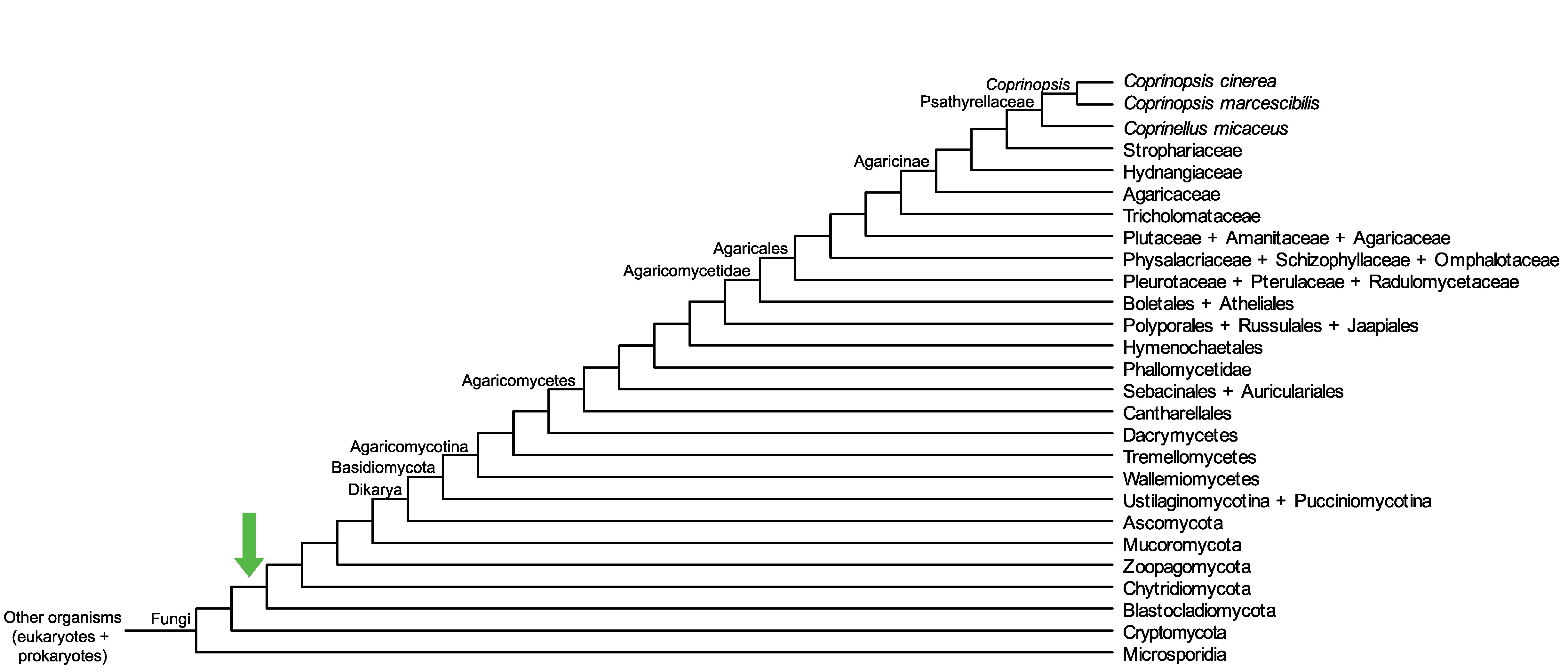
Conservation of CopciAB_539376 across fungi.
Arrow shows the origin of gene family containing CopciAB_539376.
Protein
| Sequence id | CopciAB_539376.T0 |
|---|---|
| Sequence |
>CopciAB_539376.T0 MRVEVHNALPKLEPLPPTTTRIVVTRFMIGWPQAISALTHRKPLPATTMSSPSPSSSSPLSASRGQIHVKLISAR ALNVRSVHARPYVVVQFEQNEFVSRDPTDEGDKEVKGVATAINSLSCSPSTPSISRDTSSSALSALGAIHAKASA AAAKAGRPVVQKTPSTTSIHLHPPSAPSVRTSSTSSTSSNTSTNAGGLFSRLSAHNPVWKHEVSFDVTSEDSQIT LNVYDRAASDQGFLGTVQIKPILVHDHTVDQWFKLKPYENELVTGEIRVQITYEQYKTKRALTPRDFEFLKLIGR GTFGKVFQVRKKDTKRIYAMKVLSKKEIIAKKEVAHTIGERKILQYSLSCPFLVGLKFSFQTDMDLYLVTDFKSG GELFWHLQRETRFSEERARFYVAELILALEHLHKFNIVYRDLKPENILLDATGHVALCDFGLSKANLRADELTTT FCGTTEYLAPEILLDEQGYSKIVDFWSLGVLLFEMCCGWSPFYAEDVQQMYKNICFGKIRFPKGVIGEDGKQFVK GLLNRNPKHRLGAQRDAEELKEHPFFSCIDWTALSLKQVTPPFKPVVESDESTNNFDPEFTEADIRDPKLAGMFL DDEDDHDDLGYSSDKGGKEWLDEDDPSEDWVNAYEKEKNMGHHTPNGPLGWELHANKEKNEHPPKSASIQIKPSK KAAKKHAGTPLTSSVQENFRGFTYSGGESVVASMAAHAMRNRDNDWTTQPNLNGHDSPAASHPPSRNGSVGSHDG VTFSTSKSRSKKLQDEKQEPTTEDEAYDYSTSAGRLANAKRKEVKWVDSEIDFGDF |
| Length | 806 |
Coding
| Sequence id | CopciAB_539376.T0 |
|---|---|
| Sequence |
>CopciAB_539376.T0 ATGCGAGTCGAAGTCCACAATGCTCTTCCAAAACTTGAACCTCTCCCACCAACAACAACTCGCATTGTCGTCACG CGATTCATGATTGGCTGGCCACAAGCTATCTCGGCCCTGACTCACCGAAAACCTCTCCCCGCGACCACCATGTCC TCCCCAAGCCCCTCCTCATCTTCCCCTCTCAGCGCATCCAGAGGACAGATACATGTCAAGCTCATCAGCGCACGC GCCCTCAACGTCCGCAGCGTCCATGCACGCCCTTACGTCGTCGTCCAGTTCGAGCAGAACGAATTCGTTTCCCGC GACCCCACAGACGAGGGCGACAAGGAGGTCAAGGGCGTCGCAACCGCAATCAACAGCCTCAGCTGCTCCCCGTCG ACACCCAGCATATCCAGAGACACAAGTAGCAGCGCCCTCTCAGCTCTAGGCGCAATCCATGCAAAAGCCTCAGCA GCGGCGGCCAAGGCGGGTAGGCCCGTGGTCCAGAAGACTCCAAGCACGACGTCAATCCACCTCCACCCTCCTTCG GCACCCTCAGTGCGCACATCGAGCACATCGAGCACATCCTCAAACACCAGCACAAACGCCGGTGGCCTCTTTTCT CGACTATCAGCACACAATCCCGTCTGGAAACACGAGGTCTCTTTCGATGTGACATCAGAAGACTCTCAAATCACA CTCAACGTGTACGATCGTGCAGCATCTGACCAGGGTTTCTTGGGCACCGTTCAAATCAAACCCATCCTCGTCCAC GACCACACCGTTGACCAGTGGTTCAAACTCAAACCCTACGAAAACGAGCTCGTCACAGGAGAAATTCGAGTCCAA ATCACCTATGAACAGTACAAGACCAAGCGCGCGCTCACCCCACGCGATTTCGAATTCCTCAAACTCATCGGACGA GGCACCTTTGGCAAGGTGTTCCAGGTACGCAAGAAGGACACAAAACGTATCTACGCCATGAAGGTTCTCTCCAAG AAGGAGATTATTGCCAAGAAGGAGGTGGCGCACACGATCGGCGAACGCAAGATCCTTCAATATTCCCTCTCATGT CCCTTCCTTGTCGGCCTTAAATTCAGCTTCCAAACTGATATGGACCTCTACCTCGTCACCGACTTCAAGAGCGGC GGAGAACTTTTCTGGCACCTGCAACGAGAAACCAGATTCAGCGAGGAGCGGGCTCGGTTCTATGTTGCGGAGCTC ATCTTGGCTTTGGAACACCTCCACAAATTCAACATCGTGTATCGGGATCTCAAGCCCGAAAACATCCTCCTCGAC GCCACAGGTCACGTCGCCCTGTGTGATTTCGGACTCTCGAAGGCCAACCTCAGAGCCGACGAGCTTACCACCACC TTCTGCGGTACGACCGAGTATCTTGCCCCTGAAATCCTCCTCGACGAGCAAGGGTATTCCAAGATTGTCGACTTT TGGAGTTTGGGAGTTTTGCTATTCGAGATGTGCTGTGGATGGAGTCCGTTCTACGCGGAAGATGTCCAGCAGATG TATAAAAACATCTGTTTCGGGAAGATTAGGTTCCCAAAGGGCGTCATTGGAGAGGACGGGAAGCAATTTGTCAAG GGGCTCCTCAACCGCAATCCCAAGCATCGACTGGGTGCGCAACGTGATGCTGAAGAACTCAAGGAACACCCATTC TTCTCATGCATTGACTGGACCGCCCTGTCCCTCAAGCAAGTCACTCCGCCTTTCAAGCCCGTTGTGGAGTCCGAT GAATCCACCAACAACTTTGACCCGGAGTTCACGGAAGCCGACATTCGCGACCCCAAACTCGCCGGGATGTTCCTG GACGATGAGGACGACCACGATGATCTCGGATACTCGAGCGACAAGGGCGGGAAGGAATGGCTGGACGAGGATGAT CCCAGTGAAGATTGGGTCAACGCATACGAGAAGGAGAAGAACATGGGCCACCATACCCCTAACGGCCCTCTTGGT TGGGAACTTCACGCAAACAAGGAGAAGAATGAACATCCTCCCAAATCCGCCAGCATCCAGATCAAACCTTCAAAG AAGGCGGCCAAGAAGCACGCCGGCACGCCATTGACAAGCAGTGTCCAGGAGAACTTCCGCGGATTCACCTACAGC GGCGGCGAGAGTGTGGTGGCAAGCATGGCTGCCCATGCGATGAGGAACCGAGACAACGACTGGACCACCCAACCT AACCTCAACGGCCACGATTCCCCCGCTGCCTCCCATCCTCCTTCAAGGAACGGGAGCGTCGGCAGCCACGACGGG GTCACATTCTCGACAAGTAAATCACGGTCGAAGAAGCTCCAGGACGAGAAACAAGAGCCAACGACGGAGGACGAG GCATATGACTATAGTACCAGCGCCGGCCGCCTCGCCAATGCGAAGCGGAAGGAGGTCAAGTGGGTGGACAGCGAG ATTGATTTTGGGGATTTCTAA |
| Length | 2421 |
Transcript
| Sequence id | CopciAB_539376.T0 |
|---|---|
| Sequence |
>CopciAB_539376.T0 AATTTTTCCCGCTCCAATGCGAGTCGAAGTCCACAATGCTCTTCCAAAACTTGAACCTCTCCCACCAACAACAAC TCGCATTGTCGTCACGCGATTCATGATTGGCTGGCCACAAGCTATCTCGGCCCTGACTCACCGAAAACCTCTCCC CGCGACCACCATGTCCTCCCCAAGCCCCTCCTCATCTTCCCCTCTCAGCGCATCCAGAGGACAGATACATGTCAA GCTCATCAGCGCACGCGCCCTCAACGTCCGCAGCGTCCATGCACGCCCTTACGTCGTCGTCCAGTTCGAGCAGAA CGAATTCGTTTCCCGCGACCCCACAGACGAGGGCGACAAGGAGGTCAAGGGCGTCGCAACCGCAATCAACAGCCT CAGCTGCTCCCCGTCGACACCCAGCATATCCAGAGACACAAGTAGCAGCGCCCTCTCAGCTCTAGGCGCAATCCA TGCAAAAGCCTCAGCAGCGGCGGCCAAGGCGGGTAGGCCCGTGGTCCAGAAGACTCCAAGCACGACGTCAATCCA CCTCCACCCTCCTTCGGCACCCTCAGTGCGCACATCGAGCACATCGAGCACATCCTCAAACACCAGCACAAACGC CGGTGGCCTCTTTTCTCGACTATCAGCACACAATCCCGTCTGGAAACACGAGGTCTCTTTCGATGTGACATCAGA AGACTCTCAAATCACACTCAACGTGTACGATCGTGCAGCATCTGACCAGGGTTTCTTGGGCACCGTTCAAATCAA ACCCATCCTCGTCCACGACCACACCGTTGACCAGTGGTTCAAACTCAAACCCTACGAAAACGAGCTCGTCACAGG AGAAATTCGAGTCCAAATCACCTATGAACAGTACAAGACCAAGCGCGCGCTCACCCCACGCGATTTCGAATTCCT CAAACTCATCGGACGAGGCACCTTTGGCAAGGTGTTCCAGGTACGCAAGAAGGACACAAAACGTATCTACGCCAT GAAGGTTCTCTCCAAGAAGGAGATTATTGCCAAGAAGGAGGTGGCGCACACGATCGGCGAACGCAAGATCCTTCA ATATTCCCTCTCATGTCCCTTCCTTGTCGGCCTTAAATTCAGCTTCCAAACTGATATGGACCTCTACCTCGTCAC CGACTTCAAGAGCGGCGGAGAACTTTTCTGGCACCTGCAACGAGAAACCAGATTCAGCGAGGAGCGGGCTCGGTT CTATGTTGCGGAGCTCATCTTGGCTTTGGAACACCTCCACAAATTCAACATCGTGTATCGGGATCTCAAGCCCGA AAACATCCTCCTCGACGCCACAGGTCACGTCGCCCTGTGTGATTTCGGACTCTCGAAGGCCAACCTCAGAGCCGA CGAGCTTACCACCACCTTCTGCGGTACGACCGAGTATCTTGCCCCTGAAATCCTCCTCGACGAGCAAGGGTATTC CAAGATTGTCGACTTTTGGAGTTTGGGAGTTTTGCTATTCGAGATGTGCTGTGGATGGAGTCCGTTCTACGCGGA AGATGTCCAGCAGATGTATAAAAACATCTGTTTCGGGAAGATTAGGTTCCCAAAGGGCGTCATTGGAGAGGACGG GAAGCAATTTGTCAAGGGGCTCCTCAACCGCAATCCCAAGCATCGACTGGGTGCGCAACGTGATGCTGAAGAACT CAAGGAACACCCATTCTTCTCATGCATTGACTGGACCGCCCTGTCCCTCAAGCAAGTCACTCCGCCTTTCAAGCC CGTTGTGGAGTCCGATGAATCCACCAACAACTTTGACCCGGAGTTCACGGAAGCCGACATTCGCGACCCCAAACT CGCCGGGATGTTCCTGGACGATGAGGACGACCACGATGATCTCGGATACTCGAGCGACAAGGGCGGGAAGGAATG GCTGGACGAGGATGATCCCAGTGAAGATTGGGTCAACGCATACGAGAAGGAGAAGAACATGGGCCACCATACCCC TAACGGCCCTCTTGGTTGGGAACTTCACGCAAACAAGGAGAAGAATGAACATCCTCCCAAATCCGCCAGCATCCA GATCAAACCTTCAAAGAAGGCGGCCAAGAAGCACGCCGGCACGCCATTGACAAGCAGTGTCCAGGAGAACTTCCG CGGATTCACCTACAGCGGCGGCGAGAGTGTGGTGGCAAGCATGGCTGCCCATGCGATGAGGAACCGAGACAACGA CTGGACCACCCAACCTAACCTCAACGGCCACGATTCCCCCGCTGCCTCCCATCCTCCTTCAAGGAACGGGAGCGT CGGCAGCCACGACGGGGTCACATTCTCGACAAGTAAATCACGGTCGAAGAAGCTCCAGGACGAGAAACAAGAGCC AACGACGGAGGACGAGGCATATGACTATAGTACCAGCGCCGGCCGCCTCGCCAATGCGAAGCGGAAGGAGGTCAA GTGGGTGGACAGCGAGATTGATTTTGGGGATTTCTAAAAGGGCAAGTGTGTTTTGTGGCTGGCCTCTTGCTTTGT CGTCTCTCACGACAATGCGGGGGGAGGAGGATCGCGGGTGTGCGATGGGATATACGTGCATATATCCATTTTTGT CTCTCGAGGCCGTTGTTCCTTGGAGAGTCACGCCCTTGGGCTCCCTTCCCTCCCTCCGACTCACCTCGCAGTCGA CTTGTCCGTCGATTGCACTCTCTGCGAAACCCCCACCTCATCACTCTCGGGACGATATGACGTCACGAGTGCAAT ATTAACGGAGAGTTCTTCGTGACATGTCCTCATACTTCATCTCTTCATCCACTCATTACCGCATCTCGACCGCTT GGAACCGGTTACCCAATGTGTTACCCCTGTACATGGCCTTCACATTCCTAGACACAGATAGATTCAGCCTTCATT AACCGCCATACCCCGTCCCCTTGGATGCCACGGTTCCACTCACCTTCGCCAGTCGCCAGCTAGGCTAATGACGCT CTGGGCTGCTATGGCCTAGACATTCGAGTCGACCTCACTTCCGCTCACCCTGTCGTGCATCGCATGGAACTTACT GCTCCCGCACTTCACGCCAGACGACATGATCAGACCATCCATTGACACGAACACCGAACACTCGCACTTGGTCTC TCGCTTGCAATCGCATCATGCTTCATTCCTCGACCGCATAGGAAATGAACCCTGCCTCCATCCATATCATCATCA TTTTTTCTTGGGTTTTACGCTGCTTTCCTCTTCTCCCTCTTCGTATCCTTCGACACCCTCCATCCATTTCTAAGC TTGCTCGCCATTTCATTATTCCCACTATCTATGCTCTCTCTCTGCTTTTCCCCATCAATCATCGCTATGCCCCAG TCGTCATATCCTTACCACCTTACCCGATAATGACCAACCACCCTTACCCTGCTCTGTACTTTAAAACGAAGAAAG GGACTATCCCATCCTTTCCTCGCTATCCAGGGGGGAGCCTTTCCAACGGACTTCCAAGTTCCCCCTACTTCGCTA GCTTGTACATATATCCTTATCACTTGCCTCTGCTTTCCTTTTCTCTTATTACTTCACTGCTTCATTCTCGCCCCC GCTTCGCATACTCAACCGCAATGTACAGTACAGAATCGTCGCATTCAATCCGCTACCGATCACCAACTTTGAAAT ACAGAATTGAATCGTGCCGTTC |
| Length | 3622 |
Gene
| Sequence id | CopciAB_539376.T0 |
|---|---|
| Sequence |
>CopciAB_539376.T0 AATTTTTCCCGCTCCAATGCGAGTCGAAGTCCACAATGCTCTTCCAAAACTTGAACCTCTCCCACCAACAACAAC TCGCATTGTCGTCACGCGATTCATGATTGGCTGGCCACAAGCTATCTCGGCCCTGACTCACCGAAAACCTCTCCC CGCGACCACCATGTCCTCCCCAAGCCCCTCCTCATCTTCCCCTCTCAGCGCATCCAGAGGACAGATACATGTCAA GCTCATCAGCGCACGCGCCCTCAACGTCCGCAGCGTCCATGCACGCCCTTACGTCGTCGTCCAGTTCGAGCAGAA CGAATTCGTTTCCCGCGACCCCACAGACGAGGGCGACAAGGAGGTCAAGGGCGTCGCAACCGCAATCAACAGCCT CAGCTGCTCCCCGTCGACACCCAGCATATCCAGAGACACAAGTAGCAGCGCCCTCTCAGCTCTAGGCGCAATCCA TGCAAAAGCCTCAGCAGCGGCGGCCAAGGCGGGTAGGCCCGTGGTCCAGAAGACTCCAAGCACGACGTCAATCCA CCTCCACCCTCCTTCGGCACCCTCAGTGCGCACATCGAGCACATCGAGCACATCCTCAAACACCAGCACAAACGC CGGTGGCCTCTTTTCTCGACTATCAGCACACAATCCCGTCTGGAAACACGAGGTCTCTTTGTGCGTGTTACCCTG CTTTTTTGCCAGCGAAAAAGGCGTCTGACTCCACACTGCTTGCAGCGATGTGACATCAGAAGACTCTCAAATCAC ACTCAACGTGTACGATCGTGCAGCATCTGACCAGGGTTTCTTGGGCACCGTTCAAATCAAACCCATCCTCGTCCA CGACCACACCGTTGACCAGTGGTTCAAGTCGGTCCTTTATCAATTAAGCCCGATGATCCGATACTCATTCCTCTC TTCCCGCAGACTCAAACCCTACGAAAACGAGCTCGTCACAGGAGAAATTCGAGTCCAAATCACCTATGAACAGTA CAAGGTTCGTCATTCACCCGTTTGTCAGATTTCAATTGTCACCAACATCGATGCTCCACTCCATAGACCAAGCGC GCGCTCACCCCACGCGATTTCGAATTCCTCAAACTCATCGGACGAGGCACCTTTGGCAAGGTGTTCCAGGTACGC AAGAAGGACACAAAACGTATCTACGCCATGAAGGTTCTCTCCAAGAAGGAGATTATTGCCAAGAAGGAGGTGGCG CACACGATCGGCGAACGCAAGATCCTTCAATATTCCCTCTCATGTCCCTTCCTTGTCGGCCTTAAATTCAGCTTC CAAACTGATATGGACCTCTACCTCGTCACCGACTTCAAGAGCGGCGGAGAACTTTTCTGGCACCTGCAACGAGAA ACCAGATTCAGCGAGGAGCGGGCTCGGTTCTATGTTGCGGAGCTCATCTTGGCTTTGGAACACCTCCACAAATTC AACATCGTGTATCGGGATCTCAAGCCCGAAAACATCCTCCTCGACGCCACAGGTCACGTCGCCCTGTGTGATTTC GGACTCTCGAAGGCCAACCTCAGAGCCGACGAGCTTACCACCACCTTCTGCGGTACGACCGAGTATCTTGCCCCT GAAATCCTCCTCGACGAGCAAGGGTATTCCAAGATTGTCGACTTTTGGAGTTTGGGAGTTTTGCTATTCGAGATG TGCTGTGGATGGAGTCCGTTCTACGCGGAAGATGTCCAGCAGATGTATAAAAACATCTGTTTCGGGAAGATTAGG TTCCCAAAGGGCGTCATTGGAGAGGACGGGAAGCAATTTGTCAAGGGGGTCAGTTATCTTATCTATCCTCATCTT TGGGATCCACAAGCTAATTTTATGGCCAGCTCCTCAACCGCAATCCCAAGCATCGACTGGGTGCGCAACGTGATG CTGAAGAACTCAAGGAACACCCATTCTTCTCATGCATTGACTGGACCGCCCTGTCCCTCAAGCAAGTCACTCCGC CTTTCAAGCCCGTTGTGGAGTCCGATGAATCCACCAACAACTTTGACCCGGAGTTCACGGAAGCCGACATTCGCG ACCCCAAACTCGCCGGGATGTTCCTGGACGATGAGGACGACCACGATGATCTCGGATACTCGAGCGACAAGGGCG GGAAGGAATGGCTGGACGAGGATGATCCCAGTGAAGATTGGGTCAACGCATACGAGAAGGAGAAGAACATGGGCC ACCATACCCCTAACGGCCCTCTTGGTTGGGAACTTCACGCAAACAAGGAGAAGAATGAACATCCTCCCAAATCCG CCAGCATCCAGATCAAACCTTCAAAGAAGGCGGCCAAGAAGCACGCCGGCACGCCATTGACAAGCAGTGTCCAGG AGAACTTCCGCGGATTCACCTACAGCGGCGGCGAGAGTGTGGTGGCAAGCATGGCTGCCCATGCGATGAGGAACC GAGACAACGACTGGACCACCCAACCTAACCTCAACGGCCACGATTCCCCCGCTGCCTCCCATCCTCCTTCAAGGA ACGGGAGCGTCGGCAGCCACGACGGGGTCACATTCTCGACAAGTAAATCACGGTCGAAGAAGCTCCAGGACGAGA AACAAGAGCCAACGACGGAGGACGAGGCATATGACTATAGTACCAGCGCCGGCCGCCTCGCCAATGCGAAGCGGA AGGAGGTCAAGTGGGTGGACAGCGAGATTGATTTTGGGGATTTCTAAAAGGGCAAGTGTGTTTTGTGGCTGGCCT CTTGCTTTGTCGTCTCTCACGACAATGCGGGGGGAGGAGGATCGCGGGTGTGCGATGGGATATACGTGCATATAT CCATTTTTGTCTCTCGAGGCCGTTGTTCCTTGGAGAGTCACGCCCTTGGGCTCCCTTCCCTCCCTCCGACTCACC TCGCAGTCGACTTGTCCGTCGATTGCACTCTCTGCGAAACCCCCACCTCATCACTCTCGGGACGATATGACGTCA CGAGTGCAATATTAACGGAGAGTTCTTCGTGACATGTCCTCATACTTCATCTCTTCATCCACTCATTACCGCATC TCGACCGCTTGGAACCGGTTACCCAATGTGTTACCCCTGTACATGGCCTTCACATTCCTAGACACAGATAGATTC AGCCTTCATTAACCGCCATACCCCGTCCCCTTGGATGCCACGGTTCCACTCACCTTCGCCAGTCGCCAGCTAGGC TAATGACGCTCTGGGCTGCTATGGCCTAGACATTCGAGTCGACCTCACTTCCGCTCACCCTGTCGTGCATCGCAT GGAACTTACTGCTCCCGCACTTCACGCCAGACGACATGATCAGACCATCCATTGACACGAACACCGAACACTCGC ACTTGGTCTCTCGCTTGCAATCGCATCATGCTTCATTCCTCGACCGCATAGGAAATGAACCCTGCCTCCATCCAT ATCATCATCATTTTTTCTTGGGTTTTACGCTGCTTTCCTCTTCTCCCTCTTCGTATCCTTCGACACCCTCCATCC ATTTCTAAGCTTGCTCGCCATTTCATTATTCCCACTATCTATGCTCTCTCTCTGCTTTTCCCCATCAATCATCGC TATGCCCCAGTCGTCATATCCTTACCACCTTACCCGATAATGACCAACCACCCTTACCCTGCTCTGTACTTTAAA ACGAAGAAAGGGACTATCCCATCCTTTCCTCGCTATCCAGGGGGGAGCCTTTCCAACGGACTTCCAAGTTCCCCC TACTTCGCTAGCTTGTACATATATCCTTATCACTTGCCTCTGCTTTCCTTTTCTCTTATTACTTCACTGCTTCAT TCTCGCCCCCGCTTCGCATACTCAACCGCAATGTACAGTACAGAATCGTCGCATTCAATCCGCTACCGATCACCA ACTTTGAAATACAGAATTGAATCGTGCCGTTC |
| Length | 3857 |