CopciAB_543068
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_543068 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 543068 |
| Uniprot id | Functional description | kinase-like protein | |
| Location | scaffold_7:785860..789638 | Strand | + |
| Gene length (nt) | 3779 | Transcript length (nt) | 3284 |
| CDS length (nt) | 2952 | Protein length (aa) | 983 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262791 | 47.4 | 7.719E-276 | 869 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14785 | 40.2 | 5.655E-212 | 683 |
| Lentinula edodes NBRC 111202 | Lenedo1_1122895 | 39.8 | 6.093E-208 | 671 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_3438 | 39.6 | 6.41E-207 | 668 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1459245 | 36.3 | 4.895E-166 | 548 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1108217 | 35.6 | 1.234E-157 | 523 |
| Pleurotus ostreatus PC9 | PleosPC9_1_98021 | 34.6 | 1.431E-152 | 508 |
| Lentinula edodes B17 | Lened_B_1_1_15625 | 38.6 | 1.92E-139 | 469 |
| Auricularia subglabra | Aurde3_1_1249608 | 38.3 | 7.747E-49 | 191 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_543068.T0 |
| Description | kinase-like protein |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF07714 | Protein tyrosine and serine/threonine kinase | IPR001245 | 697 | 971 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR011993 | PH-like domain superfamily |
| IPR001245 | Serine-threonine/tyrosine-protein kinase, catalytic domain |
| IPR035899 | Dbl homology (DH) domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR008266 | Tyrosine-protein kinase, active site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
KEGG
| KEGG Orthology |
|---|
| K04365 |
EggNOG
| COG category | Description |
|---|---|
| T | kinase-like protein |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
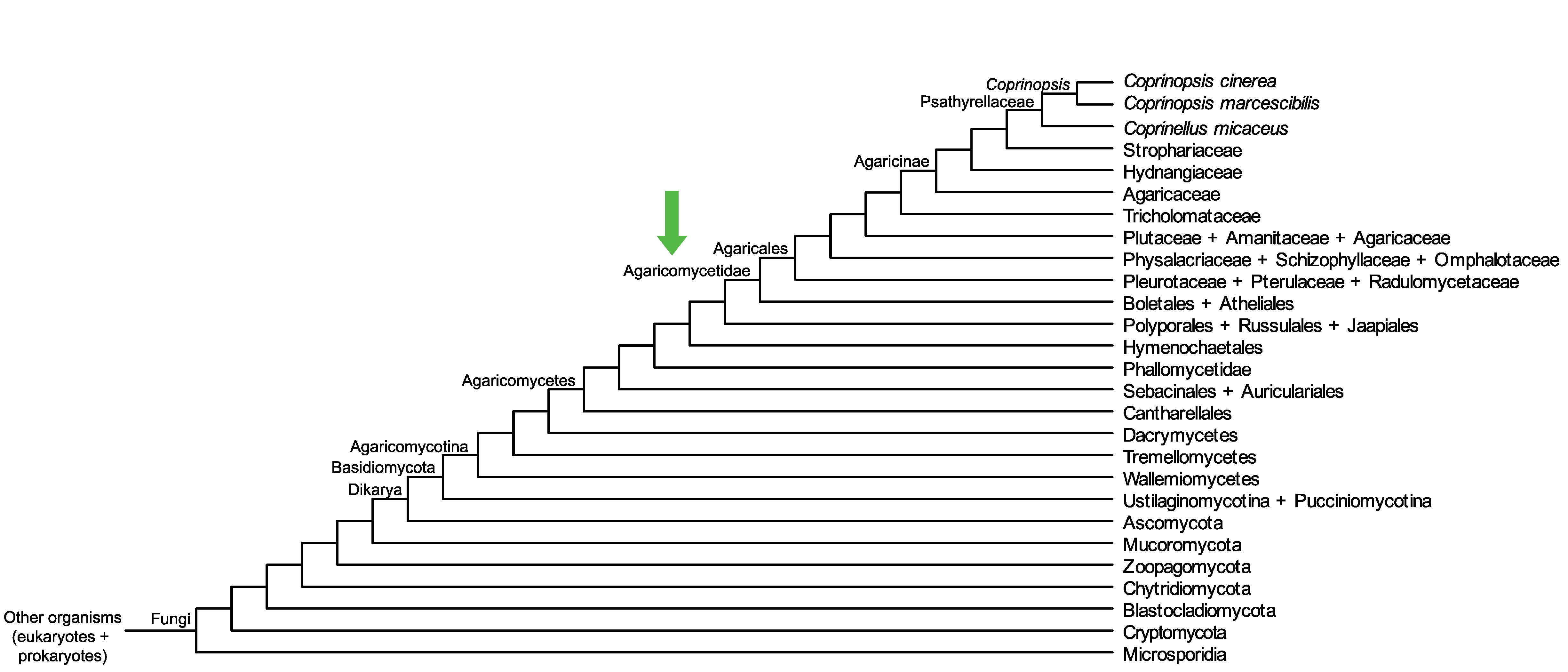
Conservation of CopciAB_543068 across fungi.
Arrow shows the origin of gene family containing CopciAB_543068.
Protein
| Sequence id | CopciAB_543068.T0 |
|---|---|
| Sequence |
>CopciAB_543068.T0 MDEHQLNGLEDVSPPTSQLSYVVFIHRVVHDTGNMPPLQPPRAPSDRVRIDSFRTLSRLQLIFEFRVWLEYLRDH VPRRYDSPMIQLWDCMSFGAPICTLLDLLGSPTPVELRVYADTFNFNLSMDERITFIESFIHRINILEIQGRIPY GEVLKVDDFISGGVPGYVRVLRTLDRILNSLHDSYPGIFVAPQGWKARRASLIQQLKEAEAIHVSRLSDVAEAAS RLYSQGDVTDPSLEGFIVNCSRLIPYHNIFYQPLGELPPPGEEIWDDIFMFSDKLLRTNIIRAYHSLCANYLPLV DTLSKLRSRHLEYADDADMVLRHLSFILGRISDYREILQAILEVSMPMNKDSYDSLCRMTFEMYQLSESIDEAGF ELRTIWAARCLKMRLGNTLRSFSSEGLGTALLDDRLVVDPVSGDHYSVFLFQSLLICCKERSSEARTADVRYPMK AWELGPALSGTCPLSIVCAVPTKNLMTIHCFDAAYFELFWVDGSNTEQSITFYPTMPQQYTQWVANLQLFVTRIL HSTSVPNMEGDDIGTGGIRTGTSFVLTAEETMYEPRRSTARAWSVIGRKGPNSESSSLVAEMGNSPGEQGSILSP NLLPTLFTPSRNGESVPASPNTPASGNNPRLPNGRNRSRTRPDFQSARSPLRSVFTRDDISPLNNAGRLGSAAPL EEGLPETLDLTGQITREGNYAVAHGGHSDVWLGRLKKQADDSDQEVKVAVKVLRNTTSDPEKKAKLVGRLQHELR VWKQLDHMHVLPLYGVTNDFGPYPSMVCPWMEKGSVSKYLEHSGDILDISERLRIIGEIMEGLQYLHLCSIIHGD LTGSNILLDTNLTAYVCDFGLSAVVLDTDTHTRASFTSHLGGAVRWTDSCFFENLDEENKEWSAPPLTYKSDVYS LGCVMLEILSGRPPYHYIALDAQVVIELHKGNKPRRPARSFVDDGQWELIQWCWTQPPEDRPDVRDVAEKLEELV AQRGDVKQ |
| Length | 983 |
Coding
| Sequence id | CopciAB_543068.T0 |
|---|---|
| Sequence |
>CopciAB_543068.T0 ATGGACGAGCACCAGCTGAATGGGTTGGAAGATGTATCGCCGCCCACATCGCAGTTGTCCTATGTTGTCTTTATC CATCGTGTAGTACACGATACAGGCAATATGCCGCCCCTCCAACCACCCAGGGCACCCTCCGACAGGGTTCGCATC GACAGTTTCCGGACTTTGTCGCGTCTTCAGCTCATATTCGAGTTTCGTGTATGGCTGGAGTATCTCCGGGACCAC GTTCCTCGCCGGTACGACTCTCCCATGATTCAGCTATGGGACTGTATGTCGTTCGGGGCACCCATTTGCACGCTC CTTGACCTCCTTGGCTCCCCAACCCCCGTCGAGCTGAGGGTGTATGCAGATACATTCAACTTTAATCTCAGCATG GACGAACGAATAACGTTCATAGAAAGCTTCATACATCGGATTAATATTCTCGAAATCCAGGGTCGAATTCCCTAT GGCGAGGTGTTAAAGGTGGATGACTTTATTAGTGGAGGTGTCCCAGGTTATGTCAGGGTGCTACGAACTCTTGAT AGGATCTTGAATTCTTTACACGACTCTTACCCTGGTATCTTTGTCGCACCTCAGGGTTGGAAAGCGCGAAGAGCG AGCTTGATCCAGCAATTGAAGGAAGCAGAGGCCATACACGTATCACGCCTTTCAGATGTTGCTGAGGCGGCATCG AGACTGTACTCCCAAGGCGACGTGACCGACCCCTCTCTGGAAGGATTCATAGTCAATTGCTCTCGCCTAATACCC TACCACAACATCTTCTATCAGCCCTTGGGCGAGCTCCCGCCTCCTGGAGAGGAGATTTGGGATGACATCTTTATG TTCTCGGACAAACTGCTACGGACCAATATCATCCGCGCATACCATTCGCTTTGTGCCAATTACCTTCCTCTTGTG GATACACTCTCTAAATTGCGCTCGAGACATCTTGAGTACGCCGACGACGCCGACATGGTCCTCCGACACCTTTCC TTTATCCTCGGCCGGATTTCAGACTACCGCGAAATTCTCCAAGCGATCCTCGAAGTGTCGATGCCCATGAACAAG GACTCCTACGACTCCCTTTGCCGAATGACCTTCGAGATGTACCAGCTGTCAGAAAGTATCGACGAGGCAGGCTTT GAATTGCGCACCATTTGGGCTGCCCGGTGTCTCAAAATGCGCTTGGGAAACACACTACGGTCTTTCAGCTCAGAA GGTCTCGGAACTGCCCTTCTCGACGATAGACTGGTAGTTGACCCAGTTTCTGGAGATCATTACTCCGTTTTCCTT TTCCAGTCTTTGCTTATCTGTTGCAAGGAGCGAAGCTCGGAGGCCAGGACTGCAGACGTACGCTATCCAATGAAG GCATGGGAACTGGGACCTGCCCTTTCGGGGACCTGTCCCTTGTCAATCGTTTGTGCTGTACCTACGAAGAACTTG ATGACGATCCATTGCTTTGACGCAGCCTATTTCGAATTATTCTGGGTGGATGGATCCAACACGGAGCAAAGCATA ACATTCTACCCGACGATGCCTCAGCAATATACACAATGGGTGGCTAATCTTCAACTCTTCGTCACTCGAATCCTG CACTCGACCTCTGTTCCGAATATGGAAGGTGACGATATTGGGACTGGAGGCATTCGTACAGGAACTTCTTTCGTC CTCACGGCGGAAGAGACGATGTACGAACCTCGCAGGTCCACAGCGCGTGCATGGAGCGTCATAGGAAGGAAAGGC CCCAATTCTGAAAGTTCCTCCCTCGTCGCGGAGATGGGGAATAGCCCGGGCGAGCAAGGTTCCATTCTGAGCCCG AACCTCTTGCCTACGCTATTCACACCCTCACGAAACGGCGAGAGCGTACCAGCAAGTCCCAACACTCCAGCCTCT GGAAATAACCCGCGTCTCCCAAACGGCCGAAACCGCTCTCGCACTCGACCCGACTTTCAATCTGCTCGAAGTCCA CTTCGATCTGTGTTTACGAGGGATGATATTTCGCCCCTCAACAATGCTGGCCGATTGGGATCCGCCGCTCCCCTG GAGGAGGGGTTACCAGAAACGCTGGATTTGACTGGACAGATCACGCGAGAAGGAAACTACGCAGTGGCTCATGGT GGTCATTCAGACGTCTGGCTGGGGAGGTTGAAGAAGCAAGCGGACGACAGCGATCAGGAGGTGAAAGTCGCTGTC AAGGTTCTCCGAAATACCACCAGCGATCCGGAGAAGAAAGCAAAACTTGTCGGTCGTCTCCAGCATGAATTGCGA GTATGGAAACAATTGGATCACATGCACGTCCTCCCCTTGTATGGTGTTACGAATGATTTCGGTCCGTATCCTTCG ATGGTGTGCCCTTGGATGGAGAAGGGGAGCGTCAGCAAATATCTGGAGCATTCCGGCGATATCCTAGATATCTCG GAACGACTGCGAATAATCGGCGAAATCATGGAAGGCCTACAATACCTCCACCTTTGCTCGATCATTCATGGGGAC TTAACAGGGTCCAACATCCTTCTCGACACCAACCTTACGGCCTATGTCTGTGACTTTGGTCTTTCTGCGGTTGTA CTCGACACCGACACTCATACGAGGGCGTCGTTTACCTCCCATTTGGGTGGGGCAGTACGGTGGACTGACTCGTGC TTCTTTGAAAACCTGGATGAGGAGAACAAAGAATGGAGCGCGCCTCCATTAACGTATAAAAGTGATGTCTATTCG CTGGGTTGTGTTATGCTCGAGATTTTGTCGGGTCGGCCACCATACCACTACATCGCGCTCGACGCTCAAGTTGTC ATCGAGTTGCACAAGGGCAACAAACCTAGGAGACCTGCTAGGTCATTCGTGGATGATGGGCAGTGGGAGTTGATT CAGTGGTGTTGGACGCAGCCGCCTGAGGATAGGCCGGATGTGCGAGATGTGGCAGAGAAACTGGAAGAATTGGTA GCACAACGAGGGGATGTGAAGCAGTAG |
| Length | 2952 |
Transcript
| Sequence id | CopciAB_543068.T0 |
|---|---|
| Sequence |
>CopciAB_543068.T0 CAACGACCACCGGCAATTGAAACGTCTGCATCGTGCAAAACTGAAACATGGCAATTATGCGAAGGCTGTACAGCT TGGAGACTTTGACAATTGTCAATGATAGGTCATTTTGATGGACGAGCACCAGCTGAATGGGTTGGAAGATGTATC GCCGCCCACATCGCAGTTGTCCTATGTTGTCTTTATCCATCGTGTAGTACACGATACAGGCAATATGCCGCCCCT CCAACCACCCAGGGCACCCTCCGACAGGGTTCGCATCGACAGTTTCCGGACTTTGTCGCGTCTTCAGCTCATATT CGAGTTTCGTGTATGGCTGGAGTATCTCCGGGACCACGTTCCTCGCCGGTACGACTCTCCCATGATTCAGCTATG GGACTGTATGTCGTTCGGGGCACCCATTTGCACGCTCCTTGACCTCCTTGGCTCCCCAACCCCCGTCGAGCTGAG GGTGTATGCAGATACATTCAACTTTAATCTCAGCATGGACGAACGAATAACGTTCATAGAAAGCTTCATACATCG GATTAATATTCTCGAAATCCAGGGTCGAATTCCCTATGGCGAGGTGTTAAAGGTGGATGACTTTATTAGTGGAGG TGTCCCAGGTTATGTCAGGGTGCTACGAACTCTTGATAGGATCTTGAATTCTTTACACGACTCTTACCCTGGTAT CTTTGTCGCACCTCAGGGTTGGAAAGCGCGAAGAGCGAGCTTGATCCAGCAATTGAAGGAAGCAGAGGCCATACA CGTATCACGCCTTTCAGATGTTGCTGAGGCGGCATCGAGACTGTACTCCCAAGGCGACGTGACCGACCCCTCTCT GGAAGGATTCATAGTCAATTGCTCTCGCCTAATACCCTACCACAACATCTTCTATCAGCCCTTGGGCGAGCTCCC GCCTCCTGGAGAGGAGATTTGGGATGACATCTTTATGTTCTCGGACAAACTGCTACGGACCAATATCATCCGCGC ATACCATTCGCTTTGTGCCAATTACCTTCCTCTTGTGGATACACTCTCTAAATTGCGCTCGAGACATCTTGAGTA CGCCGACGACGCCGACATGGTCCTCCGACACCTTTCCTTTATCCTCGGCCGGATTTCAGACTACCGCGAAATTCT CCAAGCGATCCTCGAAGTGTCGATGCCCATGAACAAGGACTCCTACGACTCCCTTTGCCGAATGACCTTCGAGAT GTACCAGCTGTCAGAAAGTATCGACGAGGCAGGCTTTGAATTGCGCACCATTTGGGCTGCCCGGTGTCTCAAAAT GCGCTTGGGAAACACACTACGGTCTTTCAGCTCAGAAGGTCTCGGAACTGCCCTTCTCGACGATAGACTGGTAGT TGACCCAGTTTCTGGAGATCATTACTCCGTTTTCCTTTTCCAGTCTTTGCTTATCTGTTGCAAGGAGCGAAGCTC GGAGGCCAGGACTGCAGACGTACGCTATCCAATGAAGGCATGGGAACTGGGACCTGCCCTTTCGGGGACCTGTCC CTTGTCAATCGTTTGTGCTGTACCTACGAAGAACTTGATGACGATCCATTGCTTTGACGCAGCCTATTTCGAATT ATTCTGGGTGGATGGATCCAACACGGAGCAAAGCATAACATTCTACCCGACGATGCCTCAGCAATATACACAATG GGTGGCTAATCTTCAACTCTTCGTCACTCGAATCCTGCACTCGACCTCTGTTCCGAATATGGAAGGTGACGATAT TGGGACTGGAGGCATTCGTACAGGAACTTCTTTCGTCCTCACGGCGGAAGAGACGATGTACGAACCTCGCAGGTC CACAGCGCGTGCATGGAGCGTCATAGGAAGGAAAGGCCCCAATTCTGAAAGTTCCTCCCTCGTCGCGGAGATGGG GAATAGCCCGGGCGAGCAAGGTTCCATTCTGAGCCCGAACCTCTTGCCTACGCTATTCACACCCTCACGAAACGG CGAGAGCGTACCAGCAAGTCCCAACACTCCAGCCTCTGGAAATAACCCGCGTCTCCCAAACGGCCGAAACCGCTC TCGCACTCGACCCGACTTTCAATCTGCTCGAAGTCCACTTCGATCTGTGTTTACGAGGGATGATATTTCGCCCCT CAACAATGCTGGCCGATTGGGATCCGCCGCTCCCCTGGAGGAGGGGTTACCAGAAACGCTGGATTTGACTGGACA GATCACGCGAGAAGGAAACTACGCAGTGGCTCATGGTGGTCATTCAGACGTCTGGCTGGGGAGGTTGAAGAAGCA AGCGGACGACAGCGATCAGGAGGTGAAAGTCGCTGTCAAGGTTCTCCGAAATACCACCAGCGATCCGGAGAAGAA AGCAAAACTTGTCGGTCGTCTCCAGCATGAATTGCGAGTATGGAAACAATTGGATCACATGCACGTCCTCCCCTT GTATGGTGTTACGAATGATTTCGGTCCGTATCCTTCGATGGTGTGCCCTTGGATGGAGAAGGGGAGCGTCAGCAA ATATCTGGAGCATTCCGGCGATATCCTAGATATCTCGGAACGACTGCGAATAATCGGCGAAATCATGGAAGGCCT ACAATACCTCCACCTTTGCTCGATCATTCATGGGGACTTAACAGGGTCCAACATCCTTCTCGACACCAACCTTAC GGCCTATGTCTGTGACTTTGGTCTTTCTGCGGTTGTACTCGACACCGACACTCATACGAGGGCGTCGTTTACCTC CCATTTGGGTGGGGCAGTACGGTGGACTGACTCGTGCTTCTTTGAAAACCTGGATGAGGAGAACAAAGAATGGAG CGCGCCTCCATTAACGTATAAAAGTGATGTCTATTCGCTGGGTTGTGTTATGCTCGAGATTTTGTCGGGTCGGCC ACCATACCACTACATCGCGCTCGACGCTCAAGTTGTCATCGAGTTGCACAAGGGCAACAAACCTAGGAGACCTGC TAGGTCATTCGTGGATGATGGGCAGTGGGAGTTGATTCAGTGGTGTTGGACGCAGCCGCCTGAGGATAGGCCGGA TGTGCGAGATGTGGCAGAGAAACTGGAAGAATTGGTAGCACAACGAGGGGATGTGAAGCAGTAGATTGACTCGAG AGATACATGTTGTTATCCACTGGTTCTGCAATGGCCCGGTATACACGCCACAGACCTTGGGTCTCAAGGCGTTAT AGTGTAGTGGTTATCACTCGGGATTTTGACTACATACCAGCAATCTTCCCGAAACCCAGGTTCGAACCCTGGTAA CGCCTGTAAGCGTCTTCCATTTCATGTTTTTGCGCGCTTTTGGTTCCCCCGGATCGGCT |
| Length | 3284 |
Gene
| Sequence id | CopciAB_543068.T0 |
|---|---|
| Sequence |
>CopciAB_543068.T0 CAACGACCACCGGCAATTGAAACGTCTGCATCGTGCAAAACTGAAACATGGCAATTATGCGAAGGCTGTACAGCT TGGAGACTTTGACAATTGTCAATGATAGGTCATTTTGATGGACGAGCACCAGCTGAATGGGTTGGAAGATGTATC GCCGCCCACATCGCAGTTGTCCTATGTTGTCTTTATCCATCGTGTAGTACACGATACAGGCAATATGCCGCCCCT CCAACCACCCAGGGCACCCTCCGACAGGGTTCGCATCGACAGTTTCCGGACTTTGTCGCGTCTTCAGCTCATATT CGAGTTTCGTGTATGGCTGGAGTATCTCCGGGACCACGTTCCTCGCCGGTACGACTCTCCCATGATTCAGCTATG GGACTGTATGTCGTTCGGGGCACCCATTTGCACGCTCCTTGACCTCCTTGGCTCCCCAACCCCCGTCGAGCTGAG GGTGTATGCAGATACATTCAACTTTAATCTCAGCATGGACGAACGAATAACGTTCATAGAAAGCTTCATACATCG GATTAATATTCTCGAAATCCAGGGTCGAATTCCCTATGGCGAGGTGTTAAAGGTGGATGACTTTATTAGTGGAGG TGTCCCAGGTTATGTCAGGGTGAGTGATCAACTTGTATACCCAACCCCTTCGCTCGCTGATCCTTGTTCATCAGG TGCTACGAACTCTTGATAGGATCTTGAATTCTTTACACGACTCTTACCCTGGTATCTTTGTCGCACCTCAGGGTT GGAAAGCGCGAAGAGCGAGCTTGATCCAGCAATTGAAGGAAGCAGAGGCCATACACGTATCACGCCTTTCAGATG TTGCTGTAGGTCCGTTCAAAACCTTTTCGCATCCACAGGACTCAAAAAACCCTAGGAGGCGGCATCGAGACTGTA CTCCCAAGGCGACGTGACCGACCCCTCTCTGGAAGGATTCATAGTCAATTGCTCTCGCCTAATACCCTACCACAA CATCTTCTATCAGCCCTTGGGCGAGCTCCCGCCTCCTGGAGAGGAGATTTGGGATGACATCTTTATGTTCTCGGT ATGGAGCTCCGACCACCTCAAACCAAGATCCATTCTGACGGAGTGATGCAGGACAAACTGCTACGGACCAATATC ATCCGCGCATACCATTCGCTTTGTGCCAATTACCTTCCTCTTGTGGATACACTCTCTAAATTGCGCTCGAGACAT GTAAGACTCCATGCTCATTTACCGTTCTCTGTTACTAAAAGCAACTTAGCTTGAGTACGCCGACGACGCCGACAT GGTCCTCCGACACCTTTCCTTTATCCTCGGCCGGATTTCAGACTACCGCGAAATTCTCCAAGCGATCCTCGAAGT GTCGATGCCCATGAACAAGGACTCCTACGACTCCCTTTGCCGAATGACCTTCGAGATGTACCAGCTGTCAGAAAG TATCGACGAGGCAGGCTTTGAATTGCGCACCATTTGGGCTGCCCGGTGTCTCAAAATGCGCTTGGGAAACACACT ACGGTCTTTCAGCTCAGAAGGTCTCGGAACTGCCCTTCTCGACGATAGACTGGTAGTTGACCCAGTTTCTGGAGA TCATTACTCCGTTTTCCTTTTCCAGTCTTTGCTTATCTGTTGCAAGGAGCGAAGCTCGGAGGCCAGGACTGCAGA CGTACGCTATCCAATGAAGGCATGGGAACTGGGACCTGCCCTTTCGGGGACCTGTCCCTTGTCAATCGTTTGTGC TGTACCTACGAAGAACTTGATGACGATCCATTGCTTTGACGCAGGTGAGTGGAACGACCCGGTTCGCCCATTTGA ATCGTAACTAACCGCGCTTCGTGTAGCCTATTTCGAATTATTCTGGGTGGATGGATCCAACACGGAGCAAAGCAT AACATTCTACCCGACGATGCCTCAGCAATATACACAATGGGTGGCTAATCTTCAACTCTTCGTCACTCGAATCCT GCACTCGACCTCTGTTCCGAATATGGAAGGTGACGATATTGGGACTGGAGGCATTCGTACAGGAACTTCTTTCGT CCTCACGGCGGAAGAGACGATGTACGAACCTCGCAGGTCCACAGCGCGTGCATGGAGCGTCATAGGAAGGAAAGG CCCCAATTCTGAAAGTTCCTCCCTCGTCGCGGAGATGGGGAATAGCCCGGGCGAGCAAGGTTCCATTCTGAGCCC GAACCTCTTGCCTACGCTATTCACACCCTCACGAAACGGCGAGAGCGTACCAGCAAGTCCCAACACTCCAGCCTC TGGAAATAACCCGCGTCTCCCAAACGGCCGAAACCGCTCTCGCACTCGACCCGACTTTCAATCTGCTCGAAGTCC ACTTCGATCTGTGTTTACGAGGGATGATATTTCGCCCCTCAACAATGCTGGCCGATTGGGATCCGCCGCTCCCCT GGAGGAGGGGTTACCAGAAACGCTGGATTTGACTGGACAGATCACGCGAGAAGGAAACTACGCAGTGGCTCATGG TGGTCATTCAGACGTCTGGCTGGGGAGGTTGAAGAAGCAAGCGGACGACAGCGATCAGGAGGTGAAAGTCGCTGT CAAGGTTCTCCGAAATACCACCAGCGATCCGGAGAAGAAAGCAAAACTTGTCGGTGTGAGTATCCGTACTGTTAC TACTTGTGCGATATGGCTAACAACCTTCTTAGCGTCTCCAGCATGAATTGCGAGTATGGAAACAATTGGATCACA TGCACGTCCTCCCCTTGTATGGTGTTACGAATGATTTCGGTCCGTATCCTTCGATGGTGTGCCCTTGGATGGAGA AGGGGAGCGTCAGCAAATATCTGGAGCATTCCGGCGATATCCTAGATATCTCGGAACGACTGCGAATAGTGAGTC GTCTCAGCTTTAGACAACTCGTACCGTTGCCATTCTGACCCCGGGGGATTTTAGATCGGCGAAATCATGGAAGGC CTACAATACCTCCACCTTTGCTCGATCATTCATGGGGACTTAACAGGGGTACGTATTCACCTCCTTCCCTTCATG GTGGCCTGGAAAGCCTGACCTATCGCCTTCAGTCCAACATCCTTCTCGACACCAACCTTACGGCCTATGTCTGTG ACTTTGGTCTTTCTGCGGTTGTACTCGACACCGACACTCATACGAGGGCGTCGTTTACCTCCCATTTGGGTGGGG CAGTACGGTGGACTGACTCGTGCTTCTTTGAAAACCTGGATGAGGAGAACAAAGAATGGAGCGCGCCTCCATTAA CGTATAAAAGTGATGTCTATTCGCTGGGTTGTGTTATGCTCGAGGTACCCTGTCTTATGCTTGGAATCTACAAAA GAGAAAAAACTCAAACAATACTCTGCAGATTTTGTCGGGTCGGCCACCATACCACTACATCGCGCTCGACGCTCA AGTTGTCATCGAGTTGCACAAGGGCAACAAACCTAGGAGACCTGCTAGGTCATTCGTGGATGATGGGCAGTGGGA GTTGATTCAGTGGTGTTGGACGCAGCCGCCTGAGGATAGGCCGGATGTGCGAGATGTGGCAGAGAAACTGGAAGA ATTGGTAGCACAACGAGGGGATGTGAAGCAGTAGATTGACTCGAGAGATACATGTTGTTATCCACTGGTTCTGCA ATGGCCCGGTATACACGCCACAGACCTTGGGTCTCAAGGCGTTATAGTGTAGTGGTTATCACTCGGGATTTTGAC TACATACCAGCAATCTTCCCGAAACCCAGGTTCGAACCCTGGTAACGCCTGTAAGCGTCTTCCATTTCATGTTTT TGCGCGCTTTTGGTTCCCCCGGATCGGCT |
| Length | 3779 |