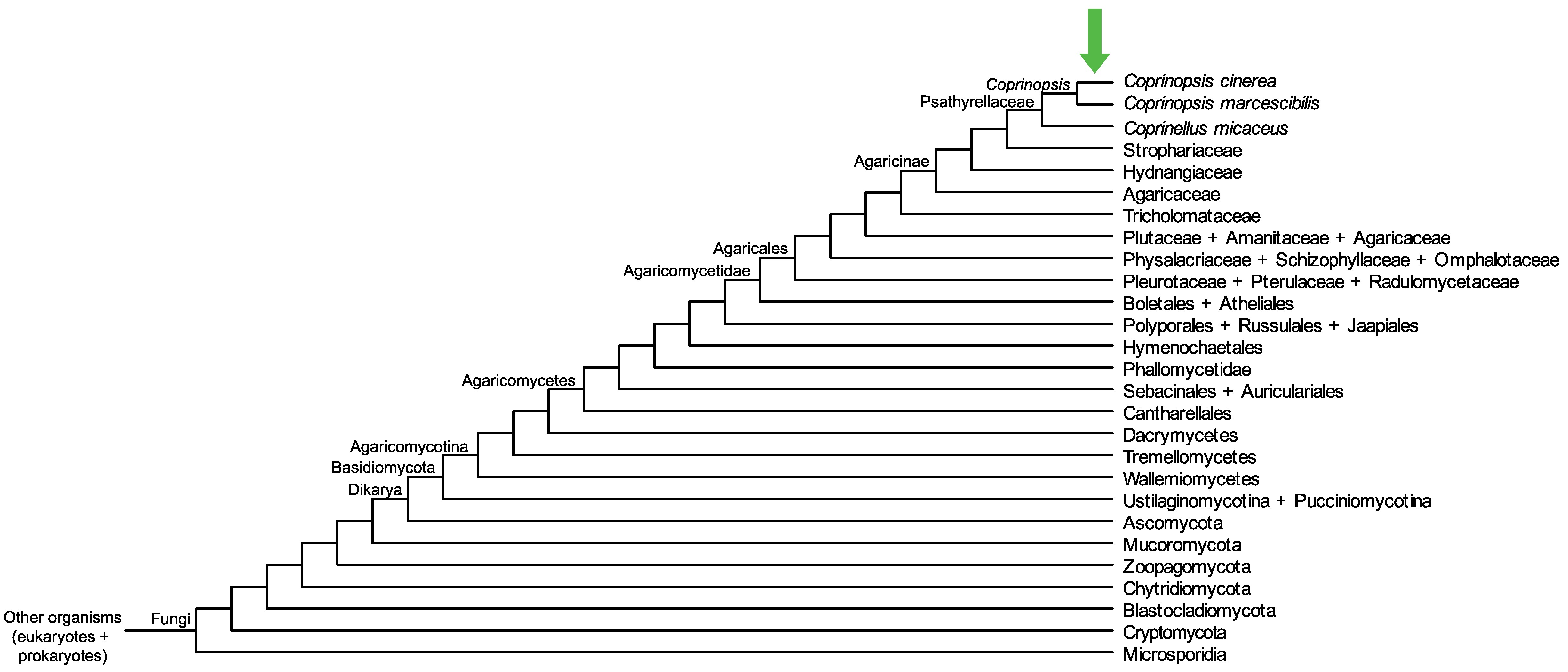
|
|
Carbon metabolism |
Transcriptional changes along alternative morphogenetic paths of C. cinerea |
Xie et al. 2020
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| sclerotia |
1.1004 |
0.3078 |
0.1777 |
| oidia |
0.9479 |
0.3709 |
0.2142 |
| vegetative_mycelium |
0.9885 |
0.554 |
0.3199 |
| primordia |
0.5461 |
0.1701 |
0.0982 |
| hyphal_knot |
1.7152 |
0.5812 |
0.3356 |
| young_fruiting_body |
0.4827 |
0.4542 |
0.2622 |
|
|
|
Complex carbon sources 1 (long incubation) |
Expression profiling on diverse complex carbon sources. |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Control - glucose |
1.2591 |
0.0802 |
0.0463 |
| Wheat bran |
2.9363 |
1.1704 |
0.6757 |
| Horse manure |
0.9093 |
0.2647 |
0.1528 |
| Oak leaves |
0.8105 |
0.4333 |
0.2502 |
| Hay |
1.5942 |
0.5261 |
0.3037 |
| Corn stalk |
0.8986 |
0.6489 |
0.3747 |
| Pectine |
0.7607 |
0.2603 |
0.1503 |
| Microcrystalline cellulose |
0.8244 |
0.1819 |
0.105 |
| Xylose |
0.8812 |
0.1689 |
0.0975 |
| Cellobiose |
1.2972 |
0.3853 |
0.2225 |
| Lignin |
0.5656 |
0.3676 |
0.2122 |
| Apple peels |
1.3617 |
0.2235 |
0.1291 |
|
|
|
Coprinopsis challenged with bacteria |
Transcriptional response to Bacillus subtilis and Escherichia coli. |
Kombrink et al. 2018
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Control |
1.2097 |
0.328 |
0.1894 |
| Bsubtilis |
0.9638 |
0.2729 |
0.1575 |
| Ecoli |
0.509 |
0.2939 |
0.1697 |
|
|
|
Early development 1 |
Time series expression profiling of basidiospore and oidium germination |
Hegedus et al.
|
QuantSeq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| BS 0h |
0.5283 |
0.3448 |
0.1991 |
| BS 4h |
0.507 |
0.3025 |
0.1747 |
| BS 8h |
0.354 |
0.1557 |
0.0899 |
| BS 12h |
0.7598 |
0.4585 |
0.2647 |
| Oidia 0 h |
0.1687 |
0.1987 |
0.1147 |
| Oidia 18 h |
0.7932 |
0.8703 |
0.5025 |
| Sclerotia |
2.4832 |
0.4742 |
0.2738 |
|
|
|
Early development 2 |
Time series expression profiling of mycelial growth, light induction and fruiting body initiation. |
Hegedus et al.
|
QuantSeq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| DM 12h |
1.3089 |
0.7999 |
0.4618 |
| DM 36h |
0.5978 |
0.3123 |
0.1803 |
| DM 60h |
1.0635 |
0.3896 |
0.225 |
| DM 84h |
2.1733 |
0.9756 |
0.5632 |
| DM 108h |
1.3489 |
0.8902 |
0.514 |
| DM 132h |
1.1755 |
0.3924 |
0.2265 |
| DM 156hAE |
2.688 |
0.9353 |
0.54 |
| ELI1hAE |
0.2686 |
0.1855 |
0.1071 |
| ELI2hAE |
0.1588 |
0.1386 |
0.08 |
| LI2hAE |
0.2928 |
0.255 |
0.1472 |
| LI6hAE |
1.4514 |
0.5475 |
0.3161 |
| LI12hAE |
1.3239 |
0.873 |
0.504 |
| LI18hAE |
1.309 |
1.2706 |
0.7336 |
| LI 24hAE |
2.2155 |
1.0291 |
0.5942 |
| DM 156hAM |
1.7497 |
0.5233 |
0.3021 |
| ELI1hAM |
0.2471 |
0.187 |
0.108 |
| ELI2hAM |
0.0982 |
0.1702 |
0.0982 |
| LI2hAM |
0.3216 |
0.3856 |
0.2226 |
| LI6hAM |
0.3275 |
0.4922 |
0.2842 |
| LI12hAM |
1.0023 |
0.6484 |
0.3743 |
| LI18hAM |
0.5533 |
0.1758 |
0.1015 |
| LI 24hAM |
2.7132 |
1.1116 |
0.6418 |
| LI 24hHK |
1.2178 |
0.5672 |
0.3275 |
| L/D 6h |
0.6412 |
0.0082 |
0.0047 |
| L/D 12h |
1.2068 |
0.3032 |
0.175 |
| L/D 18h |
0.9569 |
0.596 |
0.3441 |
| L/D 24h |
0.1907 |
0.1676 |
0.0968 |
|
|
|
Fruiting body development |
RNA-Seq analysis of fruiting body development |
Krizsan et al. 2019
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Vegetative mycelium |
2.5304 |
0.395 |
0.2281 |
| Hyphal knots |
1.7392 |
1.2278 |
0.7089 |
| Primordium 1 |
1.1842 |
0.211 |
0.1218 |
| Primordium 2 |
1.1343 |
0.2169 |
0.1252 |
| Young Fruiting body cap |
0.2787 |
0.0724 |
0.0418 |
| Younf fruiting body gill |
0.8299 |
0.4553 |
0.2629 |
| Young fruiting body stipe |
0.1083 |
0.0978 |
0.0565 |
| Mature fruiting body cap_gill |
0.5512 |
0.5033 |
0.2906 |
| Mature fruiting body stipe |
0.5474 |
0.2035 |
0.1175 |
|
|
|
Nitrogen sources (12h incubation) |
Expression profiling on diverse nitrogen sources. |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Complete medium - control |
1.3212 |
0.2572 |
0.1485 |
| NH4NO3 |
1.0838 |
0.1508 |
0.0871 |
| Proline |
0.788 |
0.2459 |
0.142 |
| Tryptophan |
0.4752 |
0.1337 |
0.0772 |
| Isoleucin |
0.6414 |
0.3669 |
0.2118 |
| Arginine |
0.4356 |
0.2343 |
0.1353 |
| Metionine |
0.3967 |
0.0506 |
0.0292 |
| NaNO3 |
0.48 |
0.0909 |
0.0525 |
|
|
|
Plant biomass (12h incubation) |
Expression profiling on diverse plant biomasses as carbon sources. |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Glucose - control |
1.3212 |
0.2572 |
0.1485 |
| Citrus peel |
0.4336 |
0.2238 |
0.1292 |
| Poplar leaves |
1.1789 |
0.4473 |
0.2582 |
| Typha leaves |
0.7461 |
0.1407 |
0.0813 |
| Overwintered Salix leaves |
0.9143 |
0.3991 |
0.2304 |
| Reed flowering |
0.9396 |
0.388 |
0.224 |
| Tigernut |
0.9266 |
0.0387 |
0.0224 |
| Energy cane |
1.0474 |
0.1312 |
0.0758 |
| Guar gum |
0.8316 |
0.1432 |
0.0827 |
| Apple peels |
0.8018 |
0.156 |
0.0901 |
| Cellobiose |
0.5817 |
0.2289 |
0.1321 |
| Corn stalk |
1.173 |
0.6852 |
0.3956 |
| Horse manure |
1.6473 |
0.5739 |
0.3313 |
| Lignin |
0.7161 |
1.0673 |
0.6162 |
| Microcrystalline cellulose |
0.7838 |
0.1988 |
0.1148 |
| Oak leaves |
1.1793 |
0.2289 |
0.1321 |
| Pectin esterified |
3.4112 |
0.3674 |
0.2121 |
| Poplar sawdust |
1.8169 |
1.065 |
0.6149 |
| Wheat bran |
1.6814 |
0.3778 |
0.2181 |
| Chlamydomonas reinhardtii |
1.0832 |
0.0202 |
0.0117 |
| Vertatryl alcohol |
0.0588 |
0.1018 |
0.0588 |
| Furfural |
1.0664 |
0.603 |
0.3481 |
| Autoclaved mycelium |
0.61 |
0.0634 |
0.0366 |
|
|
|
Simple and complex carbon and nitrogen sources |
Expression profiling on diverse complex carbon and nitrogen sources. |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Glucose control |
4.6093 |
0.1379 |
0.0796 |
| Ribose |
4.3531 |
0.4126 |
0.2382 |
| Mannose |
3.7173 |
0.5708 |
0.4036 |
| Fructose |
3.933 |
0.7811 |
0.451 |
| Arabinose |
3.869 |
0.2042 |
0.1179 |
| Xylose |
2.898 |
0.7485 |
0.4322 |
| Galacturonic acid |
4.8584 |
0.6207 |
0.3584 |
| Rhamnogalacturonan |
5.157 |
1.1051 |
0.638 |
| Pectin esterified |
3.7958 |
0.5366 |
0.3098 |
| Polygalacturonic acid |
4.3408 |
0.4446 |
0.2567 |
| Sodium acetate |
3.6878 |
0.5778 |
0.3336 |
| No nitrogen |
1.7556 |
0.4649 |
0.2684 |
| BSA |
3.2137 |
0.9079 |
0.5242 |
| Glutamine |
3.498 |
0.3855 |
0.2226 |
| No phosphate |
3.4358 |
0.4547 |
0.2625 |
| No carbon |
4.2447 |
0.6482 |
0.4584 |
| No sulphur |
1.8298 |
0.8713 |
0.503 |
|
|
|
Simple and complex carbon sources (12h incubation) |
Expression profiling on diverse complex carbon sources. |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Glucose - control |
1.3212 |
0.2572 |
0.1485 |
| Amylose |
1.7642 |
0.312 |
0.1802 |
| Fucose |
1.917 |
0.8771 |
0.5064 |
| Galactose |
1.568 |
0.1924 |
0.1111 |
| Lactose |
1.1122 |
0.0382 |
0.0221 |
| Maltose |
1.0106 |
0.2719 |
0.157 |
| Mannitol |
0.9196 |
0.3091 |
0.1785 |
| Rhamnose |
1.5047 |
0.6687 |
0.3861 |
| Sorbitol |
1.9711 |
0.6648 |
0.3838 |
| Trehalose |
1.6566 |
1.2171 |
0.7027 |
| Glycerol |
0.8977 |
0.2046 |
0.1181 |
| Glucuronic acid |
2.1468 |
0.0452 |
0.0261 |
| Arabinan |
2.1131 |
0.6368 |
0.3677 |
| Galactan |
1.1256 |
0.5227 |
0.3018 |
| Galactomannan |
2.3179 |
0.9607 |
0.5547 |
| Glucan |
1.1889 |
0.3045 |
0.1758 |
| Xylan |
1.6137 |
0.7127 |
0.4115 |
| Xyloglucan |
1.4827 |
0.1944 |
0.1122 |
| Arabinogalactan |
1.5786 |
0.1375 |
0.0794 |
| Pectin |
2.0315 |
1.0787 |
0.6228 |
| Rhamnogalacturonan |
1.1012 |
0.8818 |
0.5091 |
| Polygalacturonic acid |
1.3073 |
0.1445 |
0.0834 |
| Mannan |
1.5104 |
0.4121 |
0.2379 |
| Amylopectin |
1.7625 |
0.541 |
0.3124 |
| Inulin |
1.043 |
0.344 |
0.1986 |
| BSA |
0.4444 |
0.0637 |
0.0368 |
|
|
|
Starvation induced by water agar transfer |
Expression profiling on starvation induced by transfer to water agar |
Hegedus et al.
|
QuantSeq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Control |
1.0635 |
0.3896 |
0.225 |
| 2h WAT |
1.5792 |
0.1567 |
0.0905 |
| 4h WAT |
1.1084 |
0.8386 |
0.4841 |
| 8h WAT |
1.7231 |
1.0535 |
0.6082 |
| 16h WAT |
1.9973 |
0.4641 |
0.2679 |
| 24h WAT |
1.1992 |
1.0065 |
0.5811 |
| WAT hyphal knot -30h |
0.5831 |
0.4378 |
0.2528 |
| WAT hyphal knot -22h |
1.2898 |
0.5203 |
0.3004 |
| WAT hyphal knot -15h |
0.624 |
0.1059 |
0.0611 |
| WAT hyphal knot -7.5h |
1.2186 |
0.5881 |
0.3396 |
| WAT hyphal knot |
0.4999 |
0.1812 |
0.1046 |
| WAT hyphal knot +7.5h |
2.3522 |
0.9823 |
0.5672 |
|
|
|
Straw time series |
Time series expression profiling on straw as a carbon source |
Hegedus et al.
|
RNA-Seq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Glucose control |
0.5668 |
0.4135 |
0.2388 |
| Straw 3h |
2.071 |
1.1352 |
0.6554 |
| Straw 6h |
1.7473 |
1.2626 |
0.729 |
| Straw 12h |
1.2578 |
0.4206 |
0.2429 |
| Straw 24h |
1.0991 |
0.1151 |
0.0665 |
|
|
|
Stress conditions 1 |
Expression profiling under various stress conditions |
Hegedus et al.
|
QuantSeq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Flask control |
0.4178 |
0.0714 |
0.0412 |
| Frost (-20C, 30min) |
1.6937 |
0.5602 |
0.3234 |
| Heat shock (47C, 2h) |
0.211 |
0.3654 |
0.211 |
| High CO2 |
1.3887 |
0.1861 |
0.1075 |
| Drought (4% agar, 12h) |
1.0936 |
0.7363 |
0.4251 |
| Oxidative (1mM H2O2, 3h) |
0.8787 |
0.1638 |
0.0946 |
| Acidic (pH4, 3h) |
0.305 |
0.4214 |
0.2433 |
| Alkaline (pH9, 3h) |
0.7589 |
0.1217 |
0.0703 |
| Osmotic (sorbitol 2h) |
0.7463 |
0.3296 |
0.1903 |
| CongoRed (3h) |
0.6417 |
0.1792 |
0.1035 |
| Cobalt chloride control |
1.0091 |
0.1758 |
0.1015 |
| Cobalt chloride 2mM |
0.5835 |
0.0886 |
0.0511 |
|
|
|
Stress conditions 2 |
Expression profiling under various stress conditions |
Hegedus et al.
|
QuantSeq |
| Sample |
Mean cpm |
SD cpm |
Se of mean |
| Flask control |
0.6848 |
0.5262 |
0.3038 |
| Cold stimulation |
1.8526 |
1.9563 |
1.1295 |
| Scratched surface |
0.8317 |
0.3275 |
0.1891 |
| Ca2+, 200mM, 24h |
1.9542 |
1.8843 |
1.0879 |
| Cu2+, 2mM, 6h |
0.7368 |
0.7973 |
0.4603 |
| Voriconazole |
0.5213 |
0.204 |
0.1178 |
| Trichoderma interaction, early |
1.1516 |
0.6443 |
0.372 |
| Trichoderma interaction 60h |
0 |
0 |
0 |
| Cobalt chloride 2mM repeat |
0.6655 |
0.4942 |
0.2853 |
| Hypoxia |
1.1266 |
0.5172 |
0.2986 |
| Hypoxia control |
2.3705 |
1.0432 |
0.6023 |
| Protoplastation control |
1.7237 |
0.7472 |
0.5283 |
| Protoplastation 3.5h |
2.0355 |
2.8787 |
2.0355 |
| Protoplastation 1.5h |
1.1096 |
1.5693 |
1.1096 |
|