CC1G_13697
Coprinopsis cinerea Okoyama 7
General data
| Systematic name | CC1G_13697 | Strain | Coprinopsis cinerea A43mutB43mut pab1-1 #326 |
|---|---|---|---|
| Standard name | - | Synonyms | |
| Uniprot id | D6RK14 | Functional description | CAMK/CAMK-Unique protein kinase |
| Location | Chr_1:2477603..2478925 | Strand | + |
| Gene length (nt) | 1323 | Transcript length (nt) | 648 |
| CDS length (nt) | 648 | Protein length (aa) | 215 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| No records | |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28880 | 62.8 | 5.135E-188 | 588 |
| Lentinula edodes NBRC 111202 | Lenedo1_1125164 | 61 | 2.104E-178 | 560 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1485638 | 56.7 | 4.573E-169 | 533 |
| Schizophyllum commune H4-8 | Schco3_2684163 | 52.4 | 2.881E-157 | 499 |
| Grifola frondosa | Grifr_OBZ79927 | 53.7 | 2.749E-155 | 493 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_38805 | 51.4 | 1.019E-138 | 445 |
| Pleurotus ostreatus PC15 | PleosPC15_2_49595 | 62.7 | 1.031E-137 | 442 |
| Pleurotus ostreatus PC9 | PleosPC9_1_96060 | 62.7 | 1.009E-137 | 442 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_878 | 50.5 | 1.517E-134 | 433 |
| Auricularia subglabra | Aurde3_1_1405448 | 40.8 | 5.22E-103 | 342 |
| Flammulina velutipes | Flave_chr08AA01171 | 35.3 | 3.701E-45 | 171 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coprinopsis cinerea mycelia exposed to different biotic and abiotic stress conditions | Identification of a Novel Nematotoxic Protein in Coprinopsis cinerea | Plaza et al. 2016 | RNA-seq | |||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | 11248 |
| Description | CAMK/CAMK-Unique protein kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 3 | 87 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
| IPR000719 | Protein kinase domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
KEGG
| KEGG Orthology |
|---|
| K06641 |
| K08286 |
| K08794 |
| K12776 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
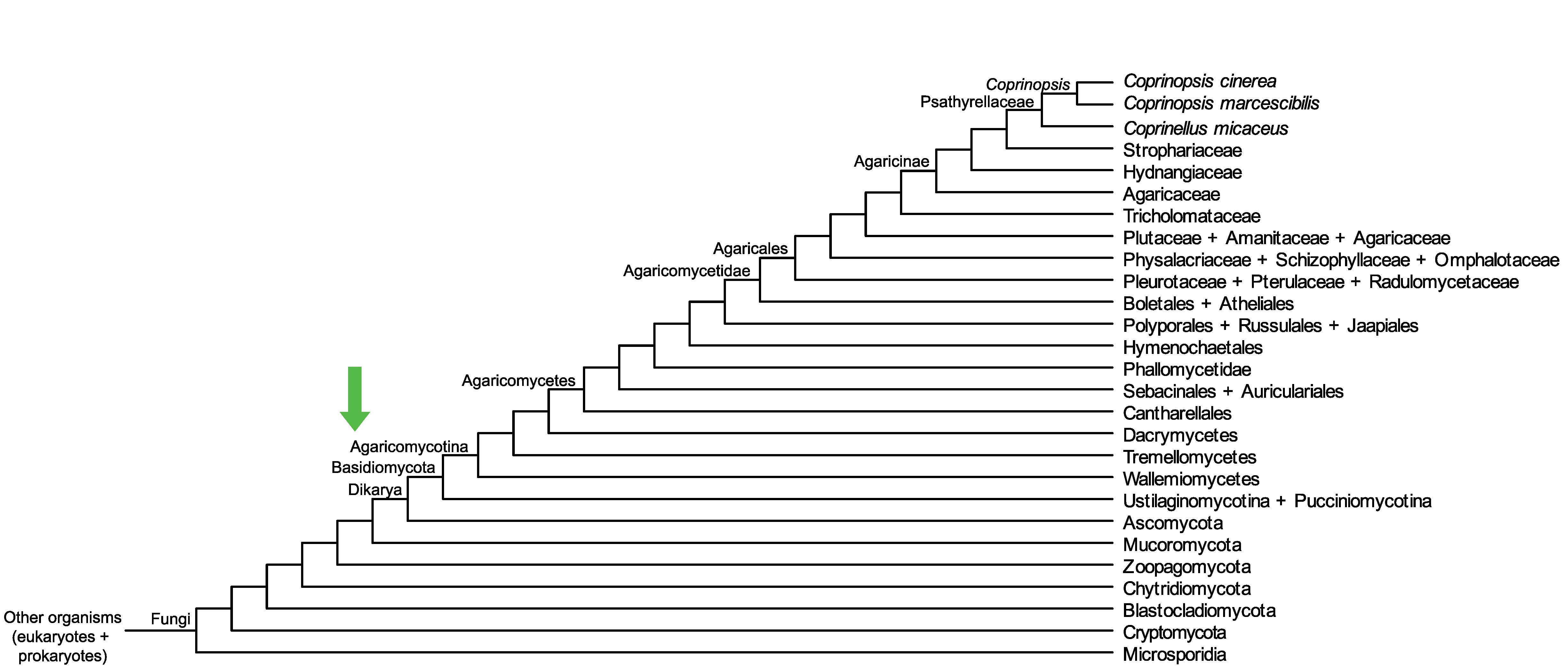
Conservation of CC1G_13697 across fungi.
Arrow shows the origin of gene family containing CC1G_13697.
Protein
| Sequence id | 11248 |
|---|---|
| Sequence |
>11248 MSQCLGTGSFATVHLAFDPTHHKQVACKSIRTRRENEVRQVLKEVRILMTLKHPNINEIYDTEEDNGFIHIFLQL CTGGDLFTYISTVNLKADRPGLCEAESNGSHPFDNDATYQSSWADDADSYTSVPTESQQCLGSFSEHRQARLKAR IVEEEVVFLHCPWSNLPQAKALVQNLLVHDYRRRANVFEALDSEWMVSEISDLQLLYRRKILGNK |
| Length | 215 |
Coding
| Sequence id | CC1G_13697T0 |
|---|---|
| Sequence |
>CC1G_13697T0 ATGTCGCAGTGCCTCGGAACGGGGTCCTTCGCCACCGTCCACCTGGCCTTCGATCCTACTCACCACAAGCAAGTA GCTTGCAAAAGCATTCGAACTCGCCGCGAGAATGAAGTTCGTCAAGTTTTGAAGGAAGTAAGGATTCTCATGACA TTGAAACATCCTAACATAAACGAGATATACGACACCGAGGAGGACAACGGATTCATTCACATCTTTCTGCAACTT TGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCTCAAAGCCGACAGGCCAGGTTTATGCGAAGCA GAGTCCAACGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCATCTTGGGCGGACGATGCGGATTCGTAT ACAAGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGCACAGACAGGCAAGACTGAAAGCGAGA ATCGTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTACCACAAGCCAAGGCATTGGTGCAGAAC TTGCTCGTCCATGATTATCGCCGGAGAGCCAACGTCTTCGAGGCACTGGACAGCGAGTGGATGGTATCGGAGATC AGCGACCTTCAGCTACTTTACCGGCGCAAGATCTTGGGCAACAAA |
| Length | 648 |
Transcript
| Sequence id | CC1G_13697T0 |
|---|---|
| Sequence |
>CC1G_13697T0 ATGTCGCAGTGCCTCGGAACGGGGTCCTTCGCCACCGTCCACCTGGCCTTCGATCCTACTCACCACAAGCAAGTA GCTTGCAAAAGCATTCGAACTCGCCGCGAGAATGAAGTTCGTCAAGTTTTGAAGGAAGTAAGGATTCTCATGACA TTGAAACATCCTAACATAAACGAGATATACGACACCGAGGAGGACAACGGATTCATTCACATCTTTCTGCAACTT TGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCTCAAAGCCGACAGGCCAGGTTTATGCGAAGCA GAGTCCAACGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCATCTTGGGCGGACGATGCGGATTCGTAT ACAAGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGCACAGACAGGCAAGACTGAAAGCGAGA ATCGTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTACCACAAGCCAAGGCATTGGTGCAGAAC TTGCTCGTCCATGATTATCGCCGGAGAGCCAACGTCTTCGAGGCACTGGACAGCGAGTGGATGGTATCGGAGATC AGCGACCTTCAGCTACTTTACCGGCGCAAGATCTTGGGCAACAAATGA |
| Length | 648 |
Gene
| Sequence id | CC1G_13697T0 |
|---|---|
| Sequence |
>CC1G_13697T0 ATGTCGCAGTGCCTCGGAACGGGGTCCTTCGCCACCGTCCACCTGGCCTTCGATCCTACTCACCACAAGCAAGTA GCTTGCAAAAGCATTCGAACTCGCCGCGAGAATGAAGTTCGTCAAGTTTTGAAGGAAGTAAGGATTCTCATGACA TTGAAACATGTAAGACTCCATTCCTATTACTCCGTGTATCTTGTTTTTCATGACGGCCTCTTAGCCTAACATAAA CGAGATATACGACACCGAGGAGGACAACGGATTCATGTATGTTTTCGTCTCCCCTCCAGAACTCTGGAATACTCA CCCAATTCGTAGTCACATCTTTCTGCAACTTTGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCT CAAAGCCGACAGGCCAGGTTTATGCGAAGCAGAGTCCAAGTATATAATGTTTCAACTCTTGACTGGGCTGGCATA CCTTCATAAGAAACATATTTCCCATCGAGGTATGCGTGTCCTTCTCACCAGTGTTCGAGGTTGCTCATTTGCCGA CTGTGTAGACTTGAAAGTGTGTTACCATCTCCACGTATGAAGCTTGGTTTACAGAATGTTGTTCCAGCCTGAAAA CATACTACTGCACGCACCGGGCCCATATCCACGGATACTAATCGCCGACTTTGGTCTCGCTCGGCCCAATGCATA TCAAGAGACGCTGAACGTATGTGGGACAGTATCGTACTTGCCTCCAGAAGGTATACTAGCATTAGACAACAAGCA CCTGGGATACATTGGGTATGGTCTCTTCCCATTCCAACCCACTGATTCAGACACTTGGTATTCACTGCAACCAGA ATGCCCGCAGACTGCTGGAGCGCTGGCGTAATCCTGTACATCATGCTGTGGTAAGCAATGTCCTACATTACTCGC GAAGAAATGTTATGGTTGACTGTCTGGACTATAGCGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCAT CTTGGGCGGACGATGCGGATTCGTATACAAGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGC ACAGACAGGCAAGACTGAAAGCGAGAATCGTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTAC CACAAGGTGGGTTTGAGTGTTCGGGTCATTCAATGAACTGACACCGTCGTTTCAGCCAAGGCATTGGTGCAGAAC TTGCTCGTCCATGATTATCGCCGGAGAGCCAACGTCTTCGAGGCACTGGACAGCGAGTGGATGGTATCGGAGATC AGCGACCTTCAGCTACTTTACCGGCGCAAGATCTTGGGCAACAAATGA |
| Length | 1323 |