CopciAB_357261
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_357261 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 357261 |
| Uniprot id | Functional description | Protein tyrosine kinase | |
| Location | scaffold_1:1830717..1832936 | Strand | - |
| Gene length (nt) | 2220 | Transcript length (nt) | 1519 |
| CDS length (nt) | 1395 | Protein length (aa) | 464 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28880 | 62.8 | 5.135E-188 | 588 |
| Lentinula edodes NBRC 111202 | Lenedo1_1125164 | 61 | 2.104E-178 | 560 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1485638 | 56.7 | 4.573E-169 | 533 |
| Schizophyllum commune H4-8 | Schco3_2684163 | 52.4 | 2.881E-157 | 499 |
| Grifola frondosa | Grifr_OBZ79927 | 53.7 | 2.749E-155 | 493 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_38805 | 51.4 | 1.019E-138 | 445 |
| Pleurotus ostreatus PC15 | PleosPC15_2_49595 | 62.7 | 1.031E-137 | 442 |
| Pleurotus ostreatus PC9 | PleosPC9_1_96060 | 62.7 | 1.009E-137 | 442 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_878 | 50.5 | 1.517E-134 | 433 |
| Auricularia subglabra | Aurde3_1_1405448 | 40.8 | 5.22E-103 | 342 |
| Flammulina velutipes | Flave_chr08AA01171 | 35.3 | 3.701E-45 | 171 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_357261.T0 |
| Description | Protein tyrosine kinase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00498 | FHA domain | IPR000253 | 38 | 107 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 152 | 445 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR000719 | Protein kinase domain |
| IPR000253 | Forkhead-associated (FHA) domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008984 | SMAD/FHA domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0005524 | ATP binding | MF |
| GO:0005515 | protein binding | MF |
KEGG
| KEGG Orthology |
|---|
| K06641 |
| K08286 |
| K08794 |
| K12776 |
EggNOG
| COG category | Description |
|---|---|
| T | Protein tyrosine kinase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
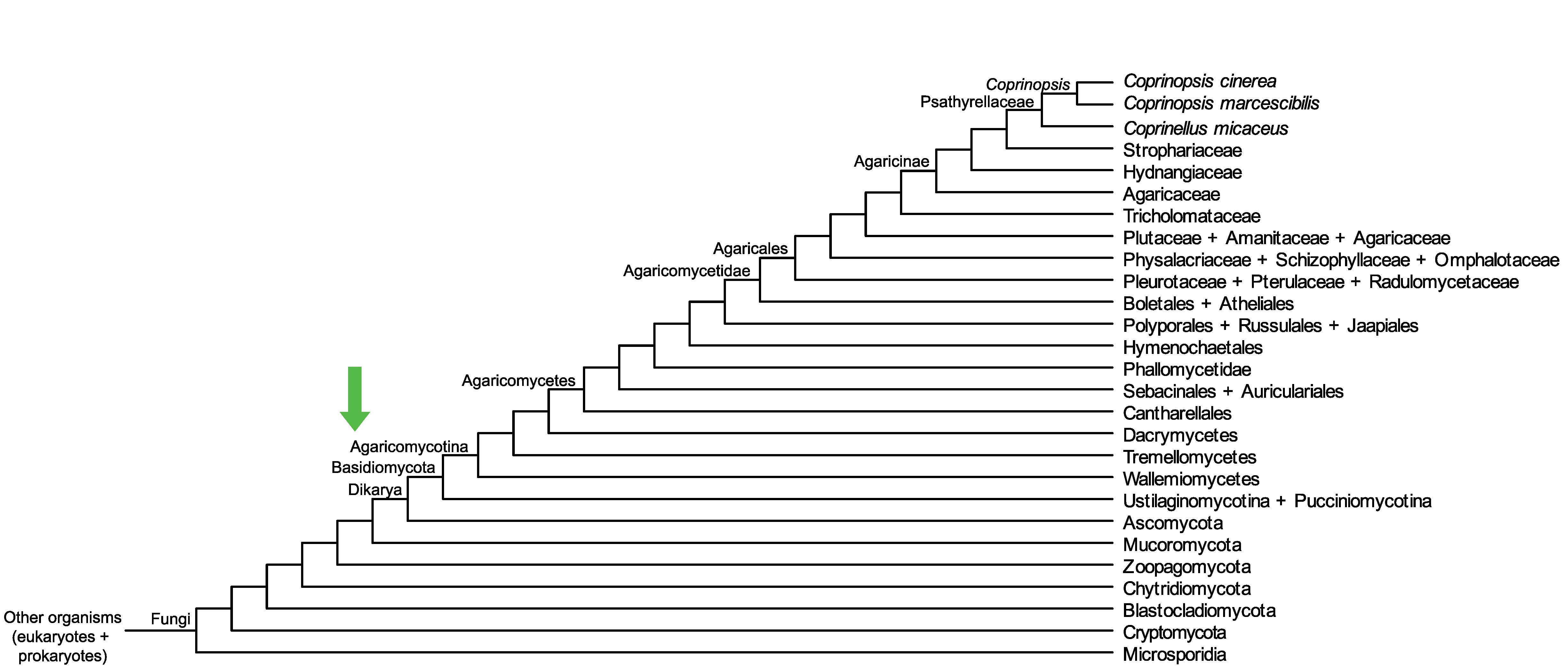
Conservation of CopciAB_357261 across fungi.
Arrow shows the origin of gene family containing CopciAB_357261.
Protein
| Sequence id | CopciAB_357261.T0 |
|---|---|
| Sequence |
>CopciAB_357261.T0 MATASEFRPSQRKDICAKLVTTRNGRKEVINLGFNCPVVIGRHPELCTYVISDSIISGVHCKLYAVPSPSGGIIV SCQDLSRNGVVLNSYRIRKASVILMDGDVLVLPNMLAFTCVHIIKGPAEKLSLFDPTPPSHARTLAQKSIGNYVV MSQCLGTGSFATVHLAFDPTHHKQVACKSIRTRRENEVRQVLKEVRILMTLKHPNINEIYDTEEDNGFIHIFLQL CTGGDLFTYISTVNLKADRPGLCEAESKYIMFQLLTGLAYLHKKHISHRDLKPENILLHAPGPYPRILIADFGLA RPNAYQETLNVCGTVSYLPPEGILALDNKHLGYIGMPADCWSAGVILYIMLCGSHPFDNDATYQSSWADDADSYT SVPTESQQCLGSFSEHRQARLKARIVEEEVVFLHCPWSNLPQAKALVQNLLVHDYRRRANVFEALDSEWMVSEIS DLQLLYRRKILGNK |
| Length | 464 |
Coding
| Sequence id | CopciAB_357261.T0 |
|---|---|
| Sequence |
>CopciAB_357261.T0 ATGGCCACCGCCTCTGAATTCCGACCCTCGCAGAGAAAAGATATATGCGCAAAGTTGGTGACGACGCGCAATGGG CGTAAGGAAGTGATCAATTTGGGTTTTAATTGCCCAGTGGTCATTGGACGGCACCCTGAGCTGTGCACCTACGTG ATCAGCGATAGCATTATCTCTGGTGTTCACTGCAAGCTATATGCAGTACCGTCGCCTTCTGGAGGGATCATTGTT TCTTGTCAAGACTTGTCACGCAATGGAGTCGTTTTGAACAGCTATCGGATACGCAAGGCTTCTGTTATTCTTATG GACGGAGACGTCCTTGTACTACCGAATATGCTGGCATTCACTTGCGTTCATATCATAAAGGGACCTGCGGAGAAG CTGTCTCTATTCGATCCAACACCTCCAAGCCACGCTAGGACGCTTGCGCAGAAGAGCATCGGGAACTATGTCGTC ATGTCGCAGTGCCTCGGAACGGGGTCCTTCGCCACCGTCCACCTGGCCTTCGATCCTACTCACCACAAGCAAGTA GCTTGCAAAAGCATTCGAACTCGCCGCGAGAATGAAGTTCGTCAAGTTTTGAAGGAAGTAAGGATTCTCATGACA TTGAAACATCCTAACATAAACGAGATATACGACACCGAGGAGGACAACGGATTCATTCACATCTTTCTGCAACTT TGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCTCAAAGCCGACAGGCCAGGTTTATGCGAAGCA GAGTCCAAGTATATAATGTTTCAACTCTTGACTGGGCTGGCATACCTTCATAAGAAACATATTTCCCATCGAGAC TTGAAACCTGAAAACATACTACTGCACGCACCGGGCCCATATCCACGGATACTAATCGCCGACTTTGGTCTCGCT CGGCCCAATGCATATCAAGAGACGCTGAACGTATGTGGGACAGTATCGTACTTGCCTCCAGAAGGTATACTAGCA TTAGACAACAAGCACCTGGGATACATTGGAATGCCCGCAGACTGCTGGAGCGCTGGCGTAATCCTGTACATCATG CTGTGCGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCATCTTGGGCGGACGATGCGGATTCGTATACA AGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGCACAGACAGGCAAGACTGAAAGCGAGAATC GTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTACCACAAGCCAAGGCATTGGTGCAGAACTTG CTCGTCCATGATTATCGCCGGAGAGCCAACGTCTTCGAGGCACTGGACAGCGAGTGGATGGTATCGGAGATCAGC GACCTTCAGCTACTTTACCGGCGCAAGATCTTGGGCAACAAATGA |
| Length | 1395 |
Transcript
| Sequence id | CopciAB_357261.T0 |
|---|---|
| Sequence |
>CopciAB_357261.T0 GTCTTTCGCTCTTGACCAGGACAATGGCCACCGCCTCTGAATTCCGACCCTCGCAGAGAAAAGATATATGCGCAA AGTTGGTGACGACGCGCAATGGGCGTAAGGAAGTGATCAATTTGGGTTTTAATTGCCCAGTGGTCATTGGACGGC ACCCTGAGCTGTGCACCTACGTGATCAGCGATAGCATTATCTCTGGTGTTCACTGCAAGCTATATGCAGTACCGT CGCCTTCTGGAGGGATCATTGTTTCTTGTCAAGACTTGTCACGCAATGGAGTCGTTTTGAACAGCTATCGGATAC GCAAGGCTTCTGTTATTCTTATGGACGGAGACGTCCTTGTACTACCGAATATGCTGGCATTCACTTGCGTTCATA TCATAAAGGGACCTGCGGAGAAGCTGTCTCTATTCGATCCAACACCTCCAAGCCACGCTAGGACGCTTGCGCAGA AGAGCATCGGGAACTATGTCGTCATGTCGCAGTGCCTCGGAACGGGGTCCTTCGCCACCGTCCACCTGGCCTTCG ATCCTACTCACCACAAGCAAGTAGCTTGCAAAAGCATTCGAACTCGCCGCGAGAATGAAGTTCGTCAAGTTTTGA AGGAAGTAAGGATTCTCATGACATTGAAACATCCTAACATAAACGAGATATACGACACCGAGGAGGACAACGGAT TCATTCACATCTTTCTGCAACTTTGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCTCAAAGCCG ACAGGCCAGGTTTATGCGAAGCAGAGTCCAAGTATATAATGTTTCAACTCTTGACTGGGCTGGCATACCTTCATA AGAAACATATTTCCCATCGAGACTTGAAACCTGAAAACATACTACTGCACGCACCGGGCCCATATCCACGGATAC TAATCGCCGACTTTGGTCTCGCTCGGCCCAATGCATATCAAGAGACGCTGAACGTATGTGGGACAGTATCGTACT TGCCTCCAGAAGGTATACTAGCATTAGACAACAAGCACCTGGGATACATTGGAATGCCCGCAGACTGCTGGAGCG CTGGCGTAATCCTGTACATCATGCTGTGCGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCATCTTGGG CGGACGATGCGGATTCGTATACAAGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGCACAGAC AGGCAAGACTGAAAGCGAGAATCGTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTACCACAAG CCAAGGCATTGGTGCAGAACTTGCTCGTCCATGATTATCGCCGGAGAGCCAACGTCTTCGAGGCACTGGACAGCG AGTGGATGGTATCGGAGATCAGCGACCTTCAGCTACTTTACCGGCGCAAGATCTTGGGCAACAAATGATTGCTTG GAAGGGTCTCTTCAAAAATATAATTATATCGTCACGTATGGAATGTCCTTCTTGATGTGTTACAAATTCATTGGT CTTCATTGTCACGGCCTCA |
| Length | 1519 |
Gene
| Sequence id | CopciAB_357261.T0 |
|---|---|
| Sequence |
>CopciAB_357261.T0 GTCTTTCGCTCTTGACCAGGACAATGGCCACCGCCTCTGAATTCCGACCCTCGCAGAGAAAAGATATATGCGCAA AGTTGGTGACGACGCGCAATGGGCGTAAGGAAGGCAAGTGGGATTTGAGCCATCCGAGCGACAGCCATTGTTAAA CGCAGCTGTGTAGTGATCAATTTGGGTTTTAATTGCCCAGTGGTCATTGGACGGCACCCTGAGCTGTGGTAAAAC TTATTCTCAGACATATATGCGCATATCTGAGCTCCCTTTTCCTAGCACCTACGTGATCAGCGATAGCATTATCTC TGGTGTTCACTGCAAGCTATATGCGTCAGTTTCCGAGAACGACAGTGACCCGGAACTGACTGAAGTCGCAGAGTA CCGTCGCCTTCTGGAGGGATCATTGTTTCTTGTCAAGTGAGTTCTATACATTGTCCTTGAAGACATTCTGTTTAC TCGTTTCAGGACTTGTCACGCAATGGAGTCGTTTTGAACAGCTATCGGATACGCAAGGCTTCTGTTATTCTTATG GACGGAGACGTCCTTGTACTACCGAATATGCTGGGTCTCAGCTATTCCCTTGCTCTTTCTGGCATCGGTCTAACT TTGTGCAGCATTCACTTGCGTTCATATCATAAAGGGACCTGCGGAGAAGCTGTCTCTATTCGATCCAACACCTCC AAGCCACGCTAGGACGCTTGCGCAGAAGGTCTTTCATCTGTTCCCGAAAGACCTGTGTTGACCCTCTTCAATATT CTGTTGGCTGACTATGGAGCTCTAGAGCATCGGGAACTATGTCGTCATGTCGCAGTGCCTCGGAACGGGGTCCTT CGCCACCGTCCACCTGGCCTTCGATCCTACTCACCACAAGCAAGTAGCTTGCAAAAGCATTCGAACTCGCCGCGA GAATGAAGTTCGTCAAGTTTTGAAGGAAGTAAGGATTCTCATGACATTGAAACATGTAAGACTCCATTCCTATTA CTCCGTGTATCTTGTTTTTCATGACGGCCTCTTAGCCTAACATAAACGAGATATACGACACCGAGGAGGACAACG GATTCATGTATGTTTTCGTCTCCCCTCCAGAACTCTGGAATACTCACCCAATTCGTAGTCACATCTTTCTGCAAC TTTGTACAGGCGGAGATTTGTTCACCTACATCTCTACCGTGAACCTCAAAGCCGACAGGCCAGGTTTATGCGAAG CAGAGTCCAAGTATATAATGTTTCAACTCTTGACTGGGCTGGCATACCTTCATAAGAAACATATTTCCCATCGAG GTATGCGTGTCCTTCTCACCAGTGTTCGAGGTTGCTCATTTGCCGACTGTGTAGACTTGAAAGTGTGTTACCATC TCCACGTATGAAGCTTGGTTTACAGAATGTTGTTCCAGCCTGAAAACATACTACTGCACGCACCGGGCCCATATC CACGGATACTAATCGCCGACTTTGGTCTCGCTCGGCCCAATGCATATCAAGAGACGCTGAACGTATGTGGGACAG TATCGTACTTGCCTCCAGAAGGTATACTAGCATTAGACAACAAGCACCTGGGATACATTGGGTATGGTCTCTTCC CATTCCAACCCACTGATTCAGACACTTGGTATTCACTGCAACCAGAATGCCCGCAGACTGCTGGAGCGCTGGCGT AATCCTGTACATCATGCTGTGGTAAGCAATGTCCTACATTACTCGCGAAGAAATGTTATGGTTGACTGTCTGGAC TATAGCGGAAGCCATCCGTTCGACAATGACGCTACCTACCAGTCATCTTGGGCGGACGATGCGGATTCGTATACA AGTGTGCCGACAGAATCACAGCAATGTCTGGGATCGTTCAGTGAGCACAGACAGGCAAGACTGAAAGCGAGAATC GTGGAAGAGGAGGTCGTCTTCTTACATTGCCCCTGGAGCAACTTACCACAAGGTGGGTTTGAGTGTTCGGGTCAT TCAATGAACTGACACCGTCGTTTCAGCCAAGGCATTGGTGCAGAACTTGCTCGTCCATGATTATCGCCGGAGAGC CAACGTCTTCGAGGCACTGGACAGCGAGTGGATGGTATCGGAGATCAGCGACCTTCAGCTACTTTACCGGCGCAA GATCTTGGGCAACAAATGATTGCTTGGAAGGGTCTCTTCAAAAATATAATTATATCGTCACGTATGGAATGTCCT TCTTGATGTGTTACAAATTCATTGGTCTTCATTGTCACGGCCTCA |
| Length | 2220 |