CopciAB_204055
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_204055 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | ATG1 | Synonyms | 204055 |
| Uniprot id | Functional description | Domain of unknown function (DUF3543) | |
| Location | scaffold_9:577703..581222 | Strand | - |
| Gene length (nt) | 3520 | Transcript length (nt) | 3100 |
| CDS length (nt) | 2523 | Protein length (aa) | 840 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17199 | 75.4 | 0 | 1313 |
| Agrocybe aegerita | Agrae_CAA7263301 | 76 | 0 | 1299 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1447819 | 70.4 | 0 | 1225 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_10401 | 71.1 | 0 | 1223 |
| Lentinula edodes NBRC 111202 | Lenedo1_1074014 | 68.7 | 0 | 1208 |
| Lentinula edodes B17 | Lened_B_1_1_11884 | 69.4 | 0 | 1172 |
| Flammulina velutipes | Flave_chr01AA00389 | 67.2 | 0 | 1123 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_120964 | 66.8 | 0 | 1118 |
| Grifola frondosa | Grifr_OBZ76318 | 62.7 | 0 | 1075 |
| Schizophyllum commune H4-8 | Schco3_2641378 | 57.2 | 6.272E-304 | 955 |
| Auricularia subglabra | Aurde3_1_1405482 | 46.8 | 2.971E-252 | 794 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1051574 | 70.2 | 3.609E-184 | 595 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_187564 | 67.5 | 2.308E-176 | 572 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_121407 | 67.2 | 1.161E-175 | 570 |
| Pleurotus ostreatus PC9 | PleosPC9_1_44366 | 79.5 | 8.633E-169 | 550 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | ATG1 |
|---|---|
| Protein id | CopciAB_204055.T0 |
| Description | Domain of unknown function (DUF3543) |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd14009 | STKc_ATG1_ULK_like | - | 26 | 315 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 22 | 315 |
| Pfam | PF12063 | Domain of unknown function (DUF3543) | IPR022708 | 562 | 788 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017441 | Protein kinase, ATP binding site |
| IPR022708 | Serine/threonine-protein kinase Atg1-like, tMIT domain |
| IPR000719 | Protein kinase domain |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR045269 | Serine/threonine-protein kinase Atg1-like |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0005524 | ATP binding | MF |
| GO:0004674 | protein serine/threonine kinase activity | MF |
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0006914 | autophagy | BP |
KEGG
| KEGG Orthology |
|---|
| K08269 |
EggNOG
| COG category | Description |
|---|---|
| O | Domain of unknown function (DUF3543) |
| T | Domain of unknown function (DUF3543) |
| U | Domain of unknown function (DUF3543) |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
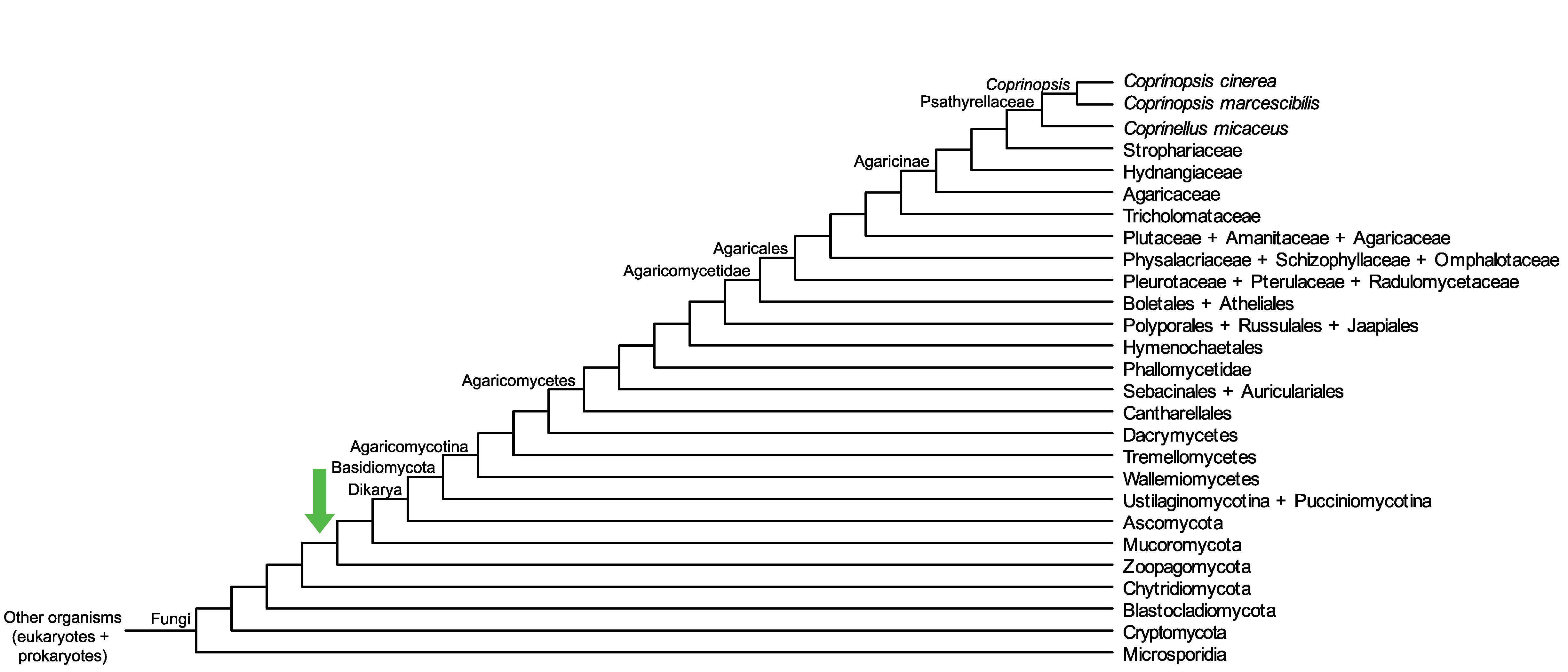
Conservation of CopciAB_204055 across fungi.
Arrow shows the origin of gene family containing CopciAB_204055.
Protein
| Sequence id | CopciAB_204055.T0 |
|---|---|
| Sequence |
>CopciAB_204055.T0 MPPIPGPSSSSNDSSDAKPYIIVSDIGRGSFATVYKGYHEETRLQVAIKAVKRDNLSARLLDNLQSEIQILKSLS HRHITKLIDIVRAEKNIYLIMEYCAGGDLTNYIKKRGRVEGLEYIPAPGEPPQYYPHPRSGGLDEIVLRSFLRQL ARALKFLRHRDLIHRDIKPQNLLLNPAPPEELARGHPLGVPILKVADFGFARSLPNAMMAETLCGSPLYMAPEIL RYEKYDAKADLWSVGAVLYEIATGRAPFRAQNHIELLKKIEQSKGLKFPDEDPKTSAEATPVPADIKKLIRALLK RNPIERASFEEFFNSTALAKSKFPRPREPVAAANAEEDHNGRPPTPDHHKDIPPEVLDPNALIPPSKFNWRRPTN GDGTEPAPNAKPVEKARRNKGLSTEGSFIPGETEEDGMLRREYVLVGDTRAVEFNRAVDEINTLPRKPLHDRKIP STPEDSPRQEYPIPNVTNPTTPHNITFPPPPNVNAPPSLSSSPSSNASRAAASALNRALSLATKKLFGTSRRQPS ISSPVHEVSSNTSPPSSPRRPQIIALDTAGERDPLEDELLANLEELAQKTDVLTRWADEMYEYVKAVPQKPLPDP TKFVKREGEGEKHARRRKLADMEAEYNAVTCVAVYMLLMSFSQKGIDKLRNFQEHMNMRHPDGDFVVSEGFDAAF SWFKDHFIKCNDRAELVKTWLPAQYDGPKAWLDQLVYDRALMLSRTAARKELLDQAAAPDECEKLYEESLWCLYA LQDDLLQTGNPFMEEDKETIGTWIKRTKLRLVRCRARMAMNDRDRIIDARADQNLADVARIPAPWDVKPSPSSTS SPTSATVRSPTSPVK |
| Length | 840 |
Coding
| Sequence id | CopciAB_204055.T0 |
|---|---|
| Sequence |
>CopciAB_204055.T0 ATGCCACCCATACCTGGCCCTTCGTCCTCGAGCAATGATTCGAGCGACGCAAAGCCCTATATCATCGTTTCTGAC ATTGGAAGGGGGAGTTTCGCTACTGTGTACAAAGGATATCATGAGGAAACGCGGTTGCAGGTCGCCATCAAGGCT GTGAAACGGGACAACCTCAGCGCCAGGCTCTTGGACAACCTGCAAAGCGAGATTCAAATCCTAAAGAGCCTGTCC CATCGACACATAACGAAGCTGATTGACATTGTGCGCGCCGAGAAGAACATCTACCTTATCATGGAGTATTGCGCG GGAGGTGACTTGACGAACTATATCAAGAAACGAGGGCGGGTAGAAGGATTGGAATACATTCCTGCACCAGGAGAA CCGCCACAATATTATCCACATCCACGGTCAGGGGGGTTGGACGAGATTGTACTGCGAAGTTTCCTCCGTCAATTA GCTCGCGCCCTCAAATTCTTGCGGCATCGAGACCTCATCCATCGTGATATCAAACCACAGAACCTGCTTCTGAAC CCGGCACCTCCAGAAGAACTGGCAAGAGGGCATCCTCTGGGTGTCCCAATCCTCAAGGTCGCCGACTTTGGTTTC GCAAGATCCCTCCCAAACGCCATGATGGCAGAGACACTCTGCGGGTCGCCCCTGTACATGGCTCCGGAAATCCTT CGCTACGAAAAGTACGACGCCAAGGCCGACCTATGGTCCGTAGGAGCAGTCCTGTACGAAATCGCCACCGGGAGA GCACCCTTCCGAGCCCAGAACCACATCGAGCTATTGAAGAAGATCGAACAATCCAAAGGGCTCAAGTTTCCTGAC GAAGACCCGAAGACGAGTGCAGAGGCGACCCCAGTTCCGGCAGACATCAAGAAGCTCATCCGAGCACTCTTGAAA CGGAATCCGATCGAGAGGGCGAGTTTTGAAGAGTTCTTTAACAGCACTGCGCTCGCCAAATCAAAGTTTCCCCGA CCTCGAGAGCCGGTTGCTGCCGCAAACGCAGAGGAAGACCACAATGGACGGCCCCCGACGCCCGATCATCACAAG GACATCCCCCCAGAAGTACTCGATCCGAACGCGCTGATCCCACCTAGCAAGTTCAATTGGAGGAGACCTACAAAC GGCGACGGGACGGAACCTGCACCAAATGCGAAACCGGTCGAAAAGGCGCGACGAAACAAGGGGCTATCAACAGAA GGTTCGTTTATCCCAGGAGAGACGGAAGAGGATGGAATGCTGCGACGAGAGTATGTGCTCGTCGGAGACACCCGA GCTGTCGAATTCAACCGTGCCGTCGACGAGATCAACACCCTTCCCCGAAAGCCCTTACACGACCGCAAGATCCCT TCCACCCCAGAGGACAGCCCACGACAAGAATACCCTATACCAAATGTCACCAACCCTACAACTCCTCACAACATC ACCTTTCCGCCTCCACCAAACGTCAACGCGCCTCCTTCATTATCGTCGTCACCATCGAGTAACGCATCGAGGGCG GCTGCAAGTGCCTTGAACAGGGCACTGTCATTAGCGACGAAGAAGCTCTTTGGAACTTCAAGACGCCAGCCGTCG ATTTCGTCCCCTGTGCATGAAGTCTCTAGCAACACAAGTCCTCCCTCTTCGCCCAGACGGCCACAGATTATAGCA CTGGACACAGCTGGCGAACGGGATCCACTGGAAGATGAGCTCTTGGCGAACCTGGAAGAACTGGCGCAGAAGACG GACGTGCTCACCCGCTGGGCGGACGAGATGTACGAATATGTGAAAGCTGTTCCCCAGAAACCACTTCCAGATCCA ACGAAGTTCGTCAAGCGCGAGGGGGAGGGGGAGAAACACGCACGAAGGCGGAAGCTGGCCGACATGGAGGCGGAA TACAACGCCGTCACCTGTGTAGCGGTTTATATGCTGCTGATGTCGTTCTCGCAGAAGGGGATCGATAAGCTGAGG AACTTCCAGGAGCATATGAATATGCGGCATCCGGATGGGGATTTCGTGGTCAGCGAAGGCTTCGATGCTGCTTTC TCGTGGTTCAAAGATCACTTCATCAAGTGTAATGACCGGGCGGAGCTCGTGAAGACATGGCTACCTGCTCAGTAC GATGGACCTAAAGCTTGGCTGGATCAGCTAGTGTATGATCGTGCTTTGATGCTGAGTCGGACAGCTGCACGAAAG GAGTTGTTGGACCAGGCCGCAGCGCCAGACGAGTGTGAGAAGCTCTACGAGGAATCGTTATGGTGCCTGTACGCC CTTCAAGATGATCTCCTGCAGACAGGTAACCCGTTCATGGAGGAAGATAAGGAAACCATTGGGACATGGATCAAG CGAACGAAGCTTCGCCTCGTTAGGTGCCGGGCGCGAATGGCCATGAACGACCGCGATCGAATAATCGATGCTCGA GCAGACCAGAACCTCGCCGACGTCGCTCGAATACCTGCTCCATGGGACGTTAAGCCATCGCCTTCATCCACCTCC TCTCCGACATCTGCCACCGTTCGATCGCCAACATCGCCTGTAAAATGA |
| Length | 2523 |
Transcript
| Sequence id | CopciAB_204055.T0 |
|---|---|
| Sequence |
>CopciAB_204055.T0 GTTCATGTTCCCTCACCTTGTAGACACTCGGTCATAGCGACTCTTGGTTTTGGAACGCTGTAGACGACCTTTTAC AAGCTGACGACGCTTGAATAGCGTTCAAGAACGACTCTGAATGCCACCCATACCTGGCCCTTCGTCCTCGAGCAA TGATTCGAGCGACGCAAAGCCCTATATCATCGTTTCTGACATTGGAAGGGGGAGTTTCGCTACTGTGTACAAAGG ATATCATGAGGAAACGCGGTTGCAGGTCGCCATCAAGGCTGTGAAACGGGACAACCTCAGCGCCAGGCTCTTGGA CAACCTGCAAAGCGAGATTCAAATCCTAAAGAGCCTGTCCCATCGACACATAACGAAGCTGATTGACATTGTGCG CGCCGAGAAGAACATCTACCTTATCATGGAGTATTGCGCGGGAGGTGACTTGACGAACTATATCAAGAAACGAGG GCGGGTAGAAGGATTGGAATACATTCCTGCACCAGGAGAACCGCCACAATATTATCCACATCCACGGTCAGGGGG GTTGGACGAGATTGTACTGCGAAGTTTCCTCCGTCAATTAGCTCGCGCCCTCAAATTCTTGCGGCATCGAGACCT CATCCATCGTGATATCAAACCACAGAACCTGCTTCTGAACCCGGCACCTCCAGAAGAACTGGCAAGAGGGCATCC TCTGGGTGTCCCAATCCTCAAGGTCGCCGACTTTGGTTTCGCAAGATCCCTCCCAAACGCCATGATGGCAGAGAC ACTCTGCGGGTCGCCCCTGTACATGGCTCCGGAAATCCTTCGCTACGAAAAGTACGACGCCAAGGCCGACCTATG GTCCGTAGGAGCAGTCCTGTACGAAATCGCCACCGGGAGAGCACCCTTCCGAGCCCAGAACCACATCGAGCTATT GAAGAAGATCGAACAATCCAAAGGGCTCAAGTTTCCTGACGAAGACCCGAAGACGAGTGCAGAGGCGACCCCAGT TCCGGCAGACATCAAGAAGCTCATCCGAGCACTCTTGAAACGGAATCCGATCGAGAGGGCGAGTTTTGAAGAGTT CTTTAACAGCACTGCGCTCGCCAAATCAAAGTTTCCCCGACCTCGAGAGCCGGTTGCTGCCGCAAACGCAGAGGA AGACCACAATGGACGGCCCCCGACGCCCGATCATCACAAGGACATCCCCCCAGAAGTACTCGATCCGAACGCGCT GATCCCACCTAGCAAGTTCAATTGGAGGAGACCTACAAACGGCGACGGGACGGAACCTGCACCAAATGCGAAACC GGTCGAAAAGGCGCGACGAAACAAGGGGCTATCAACAGAAGGTTCGTTTATCCCAGGAGAGACGGAAGAGGATGG AATGCTGCGACGAGAGTATGTGCTCGTCGGAGACACCCGAGCTGTCGAATTCAACCGTGCCGTCGACGAGATCAA CACCCTTCCCCGAAAGCCCTTACACGACCGCAAGATCCCTTCCACCCCAGAGGACAGCCCACGACAAGAATACCC TATACCAAATGTCACCAACCCTACAACTCCTCACAACATCACCTTTCCGCCTCCACCAAACGTCAACGCGCCTCC TTCATTATCGTCGTCACCATCGAGTAACGCATCGAGGGCGGCTGCAAGTGCCTTGAACAGGGCACTGTCATTAGC GACGAAGAAGCTCTTTGGAACTTCAAGACGCCAGCCGTCGATTTCGTCCCCTGTGCATGAAGTCTCTAGCAACAC AAGTCCTCCCTCTTCGCCCAGACGGCCACAGATTATAGCACTGGACACAGCTGGCGAACGGGATCCACTGGAAGA TGAGCTCTTGGCGAACCTGGAAGAACTGGCGCAGAAGACGGACGTGCTCACCCGCTGGGCGGACGAGATGTACGA ATATGTGAAAGCTGTTCCCCAGAAACCACTTCCAGATCCAACGAAGTTCGTCAAGCGCGAGGGGGAGGGGGAGAA ACACGCACGAAGGCGGAAGCTGGCCGACATGGAGGCGGAATACAACGCCGTCACCTGTGTAGCGGTTTATATGCT GCTGATGTCGTTCTCGCAGAAGGGGATCGATAAGCTGAGGAACTTCCAGGAGCATATGAATATGCGGCATCCGGA TGGGGATTTCGTGGTCAGCGAAGGCTTCGATGCTGCTTTCTCGTGGTTCAAAGATCACTTCATCAAGTGTAATGA CCGGGCGGAGCTCGTGAAGACATGGCTACCTGCTCAGTACGATGGACCTAAAGCTTGGCTGGATCAGCTAGTGTA TGATCGTGCTTTGATGCTGAGTCGGACAGCTGCACGAAAGGAGTTGTTGGACCAGGCCGCAGCGCCAGACGAGTG TGAGAAGCTCTACGAGGAATCGTTATGGTGCCTGTACGCCCTTCAAGATGATCTCCTGCAGACAGGTAACCCGTT CATGGAGGAAGATAAGGAAACCATTGGGACATGGATCAAGCGAACGAAGCTTCGCCTCGTTAGGTGCCGGGCGCG AATGGCCATGAACGACCGCGATCGAATAATCGATGCTCGAGCAGACCAGAACCTCGCCGACGTCGCTCGAATACC TGCTCCATGGGACGTTAAGCCATCGCCTTCATCCACCTCCTCTCCGACATCTGCCACCGTTCGATCGCCAACATC GCCTGTAAAATGATGCAACCAACTTTGCTATCGTTCTATTTCAAATATCTTTTCTCTCTCTTCTCCACGCTACTC ATTCATCAACAATCTGAGTTGTACCATTAGATGTATCATTATTTTTTATGAAGGCCGGGGGCACCAGGTCCGCCC GTCCGCTTTGCACGCTTCTTCACGTTCAGTTTCACGTTCACGGCCAGCATCTGTTCATTGAACACTGTCTTCACT GCAATCGCTTATATATCTATCTATTCTTAATCCCTGTTTGTTTTCATCGGTTTCCTTGTTATTTTCTTGGAGGAC ATCCCAGTTATCTTATCGTCTTGACGCAGTTGCCTGGTACGCGATGTTATCCACTTTTTTCCGTCAACCATTCTC ATTTTTCCATTCGGTGGGTTTTTACTATTTTTTCCCGAGGATGTATTTTAGTTGGTATTGTACGACCTTGTATCA TAATGCATGATGCATATTTTTGGTG |
| Length | 3100 |
Gene
| Sequence id | CopciAB_204055.T0 |
|---|---|
| Sequence |
>CopciAB_204055.T0 GTTCATGTTCCCTCACCTTGTAGACACTCGGTCATAGCGACTCTTGGTTTTGGAACGCTGTAGACGACCTTTTAC AAGCTGACGACGCTTGAATAGCGTTCAAGAACGACTCTGAATGCCACCCATACCTGGCCCTTCGTCCTCGAGCAA TGATTCGAGCGACGCAAAGCCCTATATCATCGTTTCTGACATTGGAAGGGGGAGTTTCGCTACTGTGTACAAAGG ATATCATGAGGTGTGTGCACTCTGTCGTGTAACGTATCGTAAATAACCTTTGTGCCCAGGAAACGCGGTTGCAGG TCGCCATCAAGGCTGTGAAACGGGACAACCTCAGCGCCAGGCTCTTGGACAACCTGCAAAGCGAGATTCAAATCC TAAAGAGCCTGTCCCATCGACACATAACGAAGCTGATTGACATTGTGGTACGCCAATTGATTGTGACTGACGTCC GTGACTGAGCCTTTTCCCAGCGCGCCGAGAAGAACATCTACCTTATCATGGAGTATTGCGCGGGAGGTGACTTGA CGAACTATATCAAGAAACGAGGGCGGGTAGAAGGATTGGAATACATTCCTGCACCAGGAGAACCGCCACAATATT ATCCACATCCACGGTCAGGGGGGTTGGACGAGATTGTACTGCGAAGTTTCCTCCGTCAATTAGGTACGTCGTAGC GCCCTAACCACCCGTGCCTGTGCTTATCCCTAGCTCAGCTCGCGCCCTCAAATTCTTGCGGCATCGAGACCTCAT CCATCGTGATATCAAACCACAGAACCTGCTTCTGAACCCGGCACCTCCAGAAGAACTGGCAAGAGGGCATCCTCT GGGTGTCCCAATCCTCAAGGTCGCCGACTTTGGTTTCGCAAGATCCCTCCCAAACGCCATGATGGCAGAGACACT CTGCGGGTCGCCCCTGTACATGGCTCCGGAAATCCTTCGCTACGAAAAGTACGACGCCAAGGCCGACCTATGGTC CGTAGGAGCAGTCCTGTACGAAATCGCCACCGGGAGAGCACCCTTCCGAGCCCAGAACCACATCGAGCTATTGAA GAAGATCGAACAATCCAAAGGGCTCAAGTTTCCTGACGAAGACCCGAAGACGAGTGCAGAGGCGACCCCAGTTCC GGCAGACATCAAGAAGCTCATCCGAGCACTCTTGAAACGGAATCCGATCGAGAGGGCGAGTTTTGAAGAGTTCTT TAACAGCACTGCGCTCGCCAAATCAAAGTTTCCCCGACCTCGAGAGCCGGTTGCTGCCGCAAACGCAGAGGAAGA CCACAATGGACGGCCCCCGACGCCCGATCATCACAAGGACATCCCCCCAGAAGTACTCGATCCGAACGCGCTGAT CCCACCTAGCAAGTTCAATTGGAGGAGACCTACAAACGGCGACGGGACGGAACCTGCACCAAATGCGAAACCGTA CGTATTTCATATCTGGCCGTGGAGCACTACTGACCCTCCTCTAGGGTCGAAAAGGCGCGACGAAACAAGGGGCTA TCAACAGAAGGTTCGTTTATCCCAGGAGAGACGGAAGAGGATGGAATGCTGCGACGAGAGTATGTGCTCGTCGGA GACACCCGAGCTGTCGAATTCAACCGTGCCGTCGACGAGATCAACACCCTTCCCCGAAAGCCCTTACACGACCGC AAGATCCCTTCCACCCCAGAGGACAGCCCACGACAAGAATACCCTATACCAAATGTCACCAACCCTACAACTCCT CACAACATCACCTTTCCGCCTCCACCAAACGTCAACGCGCCTCCTTCATTATCGTCGTCACCATCGAGTAACGCA TCGAGGGCGGCTGCAAGTGCCTTGAACAGGGCACTGTCATTAGCGACGAAGAAGCTCTTTGGAACTTCAAGACGC CAGCCGTCGATTTCGTCCCCTGTGCATGAAGTCTCTAGCAACACAAGTCCTCCCTCTTCGCCCAGACGGCCACAG ATTATAGCACTGGACACAGCTGGCGAACGGGATCCACTGGAAGATGAGCTCTTGGCGAACCTGGAAGAACTGGCG CAGAAGACGGACGTGCTCACCCGCTGGGCGGACGAGATGTACGAATATGTGAAAGCTGTTCCCCAGAGTACGTTG CGCTGTTCATGATCGCCATTCTTCCACTCACTTCGTATGCAGAACCACTTCCAGATCCAACGAAGTTCGTCAAGC GCGAGGGGGAGGGGGAGAAACACGCACGAAGGCGGAAGCTGGCCGACATGGAGGCGGAATACAACGCCGTCACCT GTGTAGCGGTTTATATGCTGCTGATGTCGTTCTCGCAGAAGGGGATCGATAAGCTGAGGAACTTCCAGGAGCATA TGAATATGCGGCATCCGGATGGGGATTTCGTGGTCAGCGAAGGCTTCGATGCTGGTGCGTCATTCGATCTACTCA TGAAGGTTATTATGGAGAAGTGTGATGTTGACTATGCGATTTGGGTAGCTTTCTCGTGGTTCAAAGATCACTTCA TCAAGTGTAATGACCGGGCGGAGCTCGTGAAGACATGGCTACCTGCTCAGTACGATGGACCTAAAGCTTGGCTGG ATCAGCTAGTGTATGATCGTGCTTTGATGCTGGTGAGTAGCGTCGAGGCATTTCGTTATCTCCTGATCAGTTCTG ACCTCTACACCAATAGAGTCGGACAGCTGCACGAAAGGAGTTGTTGGACCAGGCCGCAGCGCCAGACGAGTGTGA GAAGCTCTACGAGGAATCGTTATGGTGCCTGTACGCCCTTCAAGATGATCTCCTGCAGACAGGTAACCCGTTCAT GGAGGAAGATAAGGAAACCATTGGGACATGTGCGTTATTTTCTTGCTTTTGTATAATCCCAGCTGATGATGGCGC AGGGATCAAGCGAACGAAGCTTCGCCTCGTTAGGTGCCGGGCGCGAATGGCCATGAACGACCGCGATCGAATAAT CGATGCTCGAGCAGACCAGAACCTCGCCGACGTCGCTCGAATACCTGCTCCATGGGACGTTAAGCCATCGCCTTC ATCCACCTCCTCTCCGACATCTGCCACCGTTCGATCGCCAACATCGCCTGTAAAATGATGCAACCAACTTTGCTA TCGTTCTATTTCAAATATCTTTTCTCTCTCTTCTCCACGCTACTCATTCATCAACAATCTGAGTTGTACCATTAG ATGTATCATTATTTTTTATGAAGGCCGGGGGCACCAGGTCCGCCCGTCCGCTTTGCACGCTTCTTCACGTTCAGT TTCACGTTCACGGCCAGCATCTGTTCATTGAACACTGTCTTCACTGCAATCGCTTATATATCTATCTATTCTTAA TCCCTGTTTGTTTTCATCGGTTTCCTTGTTATTTTCTTGGAGGACATCCCAGTTATCTTATCGTCTTGACGCAGT TGCCTGGTACGCGATGTTATCCACTTTTTTCCGTCAACCATTCTCATTTTTCCATTCGGTGGGTTTTTACTATTT TTTCCCGAGGATGTATTTTAGTTGGTATTGTACGACCTTGTATCATAATGCATGATGCATATTTTTGGTG |
| Length | 3520 |