CopciAB_356143
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_356143 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 356143 |
| Uniprot id | Functional description | Histone-like transcription factor (CBF/NF-Y) and archaeal histone | |
| Location | scaffold_7:1424843..1425974 | Strand | - |
| Gene length (nt) | 1132 | Transcript length (nt) | 956 |
| CDS length (nt) | 738 | Protein length (aa) | 245 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Schizophyllum commune H4-8 | Schco3_2614728 | 46.5 | 3.657E-46 | 166 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB17967 | 51.8 | 1.851E-45 | 164 |
| Agrocybe aegerita | Agrae_CAA7261434 | 47.3 | 9.985E-42 | 153 |
| Lentinula edodes NBRC 111202 | Lenedo1_1043331 | 45.8 | 5.347E-39 | 145 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_12666 | 45.8 | 5.306E-39 | 145 |
| Grifola frondosa | Grifr_OBZ69003 | 45.3 | 5.246E-38 | 142 |
| Lentinula edodes B17 | Lened_B_1_1_583 | 44.1 | 2.197E-37 | 140 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_193750 | 39.7 | 6.887E-30 | 118 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_117121 | 40.5 | 8.543E-30 | 118 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_113914 | 38.8 | 4.312E-24 | 101 |
| Pleurotus ostreatus PC15 | PleosPC15_2_169408 | 41.3 | 2.56E-18 | 84 |
| Pleurotus ostreatus PC9 | PleosPC9_1_107538 | 41.3 | 5.417E-18 | 83 |
| Pleurotus eryngii ATCC 90797 | Pleery1_546724 | 42.7 | 1.406E-17 | 82 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_356143.T0 |
| Description | Histone-like transcription factor (CBF/NF-Y) and archaeal histone |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00808 | Histone-like transcription factor (CBF/NF-Y) and archaeal histone | IPR003958 | 120 | 180 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR009072 | Histone-fold |
| IPR003958 | Transcription factor CBF/NF-Y/archaeal histone domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0046982 | protein heterodimerization activity | MF |
KEGG
| KEGG Orthology |
|---|
| K03004 |
| K03506 |
EggNOG
| COG category | Description |
|---|---|
| K | Histone-like transcription factor (CBF/NF-Y) and archaeal histone |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| histone fold |
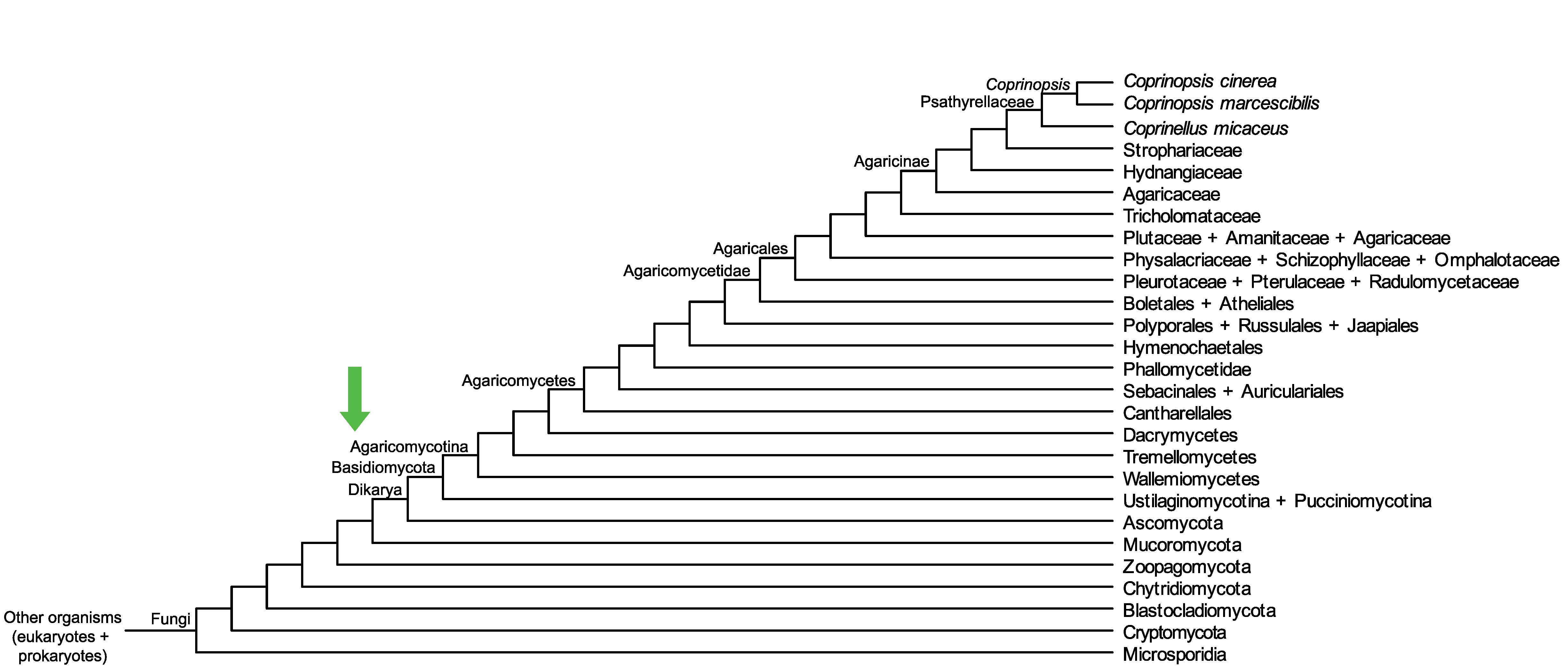
Conservation of CopciAB_356143 across fungi.
Arrow shows the origin of gene family containing CopciAB_356143.
Protein
| Sequence id | CopciAB_356143.T0 |
|---|---|
| Sequence |
>CopciAB_356143.T0 MSSSPISSPINKNHASMEEDKPMNDTLVDEEGDVVQFEDEDESQFPEEDEPQFPDEDEAQFPEEEEDAMDVDEPP PPEVEAGKGGKTKAAKTTATTSSTSKPKKKKEAQTLEREPGKSLLPLARVQKIIKADKDIPIVAKDATFLISLAT EEFIRRITEAGARVANRENRTTVQGRDIASVAKRVDEFLFLDDILPFVSAEPKRQNKVKDMISAVAPLKGAPTAL EQFVGKKGQQQESNNHEEEG |
| Length | 245 |
Coding
| Sequence id | CopciAB_356143.T0 |
|---|---|
| Sequence |
>CopciAB_356143.T0 ATGTCCTCTTCACCGATATCATCACCCATTAACAAGAATCACGCGTCAATGGAAGAGGACAAGCCCATGAACGAC ACACTGGTGGACGAGGAGGGGGATGTAGTCCAGTTTGAAGACGAAGATGAATCACAGTTTCCGGAAGAGGACGAA CCACAGTTTCCAGATGAAGATGAGGCGCAGTTTCCGGAGGAGGAGGAAGATGCGATGGATGTGGACGAACCGCCG CCTCCAGAGGTAGAGGCAGGGAAGGGCGGAAAGACCAAGGCGGCCAAGACGACGGCGACAACGAGTTCAACATCC AAACCGAAGAAGAAGAAGGAAGCGCAGACGCTTGAGAGAGAACCTGGAAAATCGTTACTCCCTCTTGCGCGTGTT CAGAAAATCATTAAAGCGGATAAAGATATACCCATAGTAGCTAAAGACGCTACGTTCCTCATTTCATTGGCAACT GAGGAGTTCATTAGACGGATAACGGAGGCTGGGGCGAGGGTCGCCAATCGAGAGAATAGGACGACTGTGCAAGGA CGGGATATAGCGAGCGTAGCGAAACGCGTGGATGAATTTCTTTTCCTTGACGATATTTTGCCTTTCGTTTCGGCA GAACCTAAACGACAAAACAAAGTAAAGGATATGATTTCAGCTGTGGCGCCGTTGAAGGGTGCTCCAACTGCTTTG GAACAGTTTGTTGGGAAGAAGGGGCAACAACAGGAAAGCAATAATCATGAGGAGGAAGGATAG |
| Length | 738 |
Transcript
| Sequence id | CopciAB_356143.T0 |
|---|---|
| Sequence |
>CopciAB_356143.T0 GGACGCGTCTCAATTCACGACCACCACGCCGCCAGGCCTTTCCTTCACATTCCTAACACACCTTGTGTATCAATC TACAACGAATCATGTCCTCTTCACCGATATCATCACCCATTAACAAGAATCACGCGTCAATGGAAGAGGACAAGC CCATGAACGACACACTGGTGGACGAGGAGGGGGATGTAGTCCAGTTTGAAGACGAAGATGAATCACAGTTTCCGG AAGAGGACGAACCACAGTTTCCAGATGAAGATGAGGCGCAGTTTCCGGAGGAGGAGGAAGATGCGATGGATGTGG ACGAACCGCCGCCTCCAGAGGTAGAGGCAGGGAAGGGCGGAAAGACCAAGGCGGCCAAGACGACGGCGACAACGA GTTCAACATCCAAACCGAAGAAGAAGAAGGAAGCGCAGACGCTTGAGAGAGAACCTGGAAAATCGTTACTCCCTC TTGCGCGTGTTCAGAAAATCATTAAAGCGGATAAAGATATACCCATAGTAGCTAAAGACGCTACGTTCCTCATTT CATTGGCAACTGAGGAGTTCATTAGACGGATAACGGAGGCTGGGGCGAGGGTCGCCAATCGAGAGAATAGGACGA CTGTGCAAGGACGGGATATAGCGAGCGTAGCGAAACGCGTGGATGAATTTCTTTTCCTTGACGATATTTTGCCTT TCGTTTCGGCAGAACCTAAACGACAAAACAAAGTAAAGGATATGATTTCAGCTGTGGCGCCGTTGAAGGGTGCTC CAACTGCTTTGGAACAGTTTGTTGGGAAGAAGGGGCAACAACAGGAAAGCAATAATCATGAGGAGGAAGGATAGC GGACCATATCGCGAGCCGCTGGAGTTCATGCAACGGCAGAGAAATTGCTATATCTAGTGTGTCGTTTCCTTTTGC ATTGAACAGTCCGAGTTGTACAGGGTCACAGAGTACATTATATATTTATGTGGATG |
| Length | 956 |
Gene
| Sequence id | CopciAB_356143.T0 |
|---|---|
| Sequence |
>CopciAB_356143.T0 GGACGCGTCTCAATTCACGACCACCACGCCGCCAGGCCTTTCCTTCACATTCCTAACACACCTTGTGTATCAATC TACAACGAATCATGTCCTCTTCACCGATATCATCACCCATTAACAAGAATCACGCGTCAATGGAAGAGGACAAGC CCATGAACGACACACTGGTGGACGAGGAGGGGGATGTAGTCCAGTTTGAAGACGAAGATGAATCACAGTTTCCGG AAGAGGACGAACCACAGTTTCCAGATGAAGATGAGGCGCAGTTTCCGGAGGAGGAGGAAGATGCGATGGATGTGG ACGAACCGCCGCCTCCAGAGGTAGAGGCAGGGAAGGGCGGAAAGACCAAGGCGGCCAAGACGACGGCGACAACGA GTTCAACATCCAAACCGAAGAAGAAGAAGGAAGCGCAGACGCTTGAGAGAGAACCTGGAAAATCGTTACTCCCTC TTGCGCGTGTTCAGAAAATCATTAAAGCGGATAAAGTGCGTTTTGGTACCGTCAGGTTCGGCGACGAGAGCTAAT GAGATTGTTTAGGATATACCCATAGTAGCTAAAGACGCTACGTTCCTCATTTCATTGGCAACTGAGGAGTTCATT AGACGGATAACGGAGGCTGGGGCGAGGGTCGCCAATCGAGAGAATAGGACGACTGTGCAAGGACGGGATATAGGT ACATTATCCTTCTCCTCCGGGTGGTGTCAGTGCTGATTATAGTATAGCGAGCGTAGCGAAACGCGTGGATGAATT TCTTTTCCTTGACGGTAAGCACTTGCAGTTGCGGTTAAGAATGAACACGGTGTCTAACGCGTTCATTGACTTCGT CTCGTTGCATATAGATATTTTGCCTTTCGTTTCGGCAGAACCTAAACGACAAAACAAAGTAAAGGATATGATTTC AGCTGTGGCGCCGTTGAAGGGTGCTCCAACTGCTTTGGAACAGTTTGTTGGGAAGAAGGGGCAACAACAGGAAAG CAATAATCATGAGGAGGAAGGATAGCGGACCATATCGCGAGCCGCTGGAGTTCATGCAACGGCAGAGAAATTGCT ATATCTAGTGTGTCGTTTCCTTTTGCATTGAACAGTCCGAGTTGTACAGGGTCACAGAGTACATTATATATTTAT GTGGATG |
| Length | 1132 |