CopciAB_357091
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_357091 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 357091 |
| Uniprot id | Functional description | Chitin synthase | |
| Location | scaffold_1:1326853..1330949 | Strand | - |
| Gene length (nt) | 4097 | Transcript length (nt) | 3172 |
| CDS length (nt) | 2547 | Protein length (aa) | 848 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Ustilago maydis | CHS2 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7264741 | 83.2 | 0 | 1458 |
| Pleurotus ostreatus PC9 | PleosPC9_1_115957 | 78.4 | 0 | 1420 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1437737 | 78 | 0 | 1416 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB28666 | 78.4 | 0 | 1409 |
| Flammulina velutipes | Flave_chr08AA00385 | 77.2 | 0 | 1398 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_114159 | 74.8 | 0 | 1368 |
| Lentinula edodes NBRC 111202 | Lenedo1_1031833 | 74.4 | 0 | 1361 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_345 | 74.5 | 0 | 1360 |
| Schizophyllum commune H4-8 | Schco3_2613031 | 73.6 | 0 | 1360 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1036823 | 83.2 | 0 | 1329 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_60607 | 78.8 | 0 | 1318 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_31788 | 78.8 | 0 | 1318 |
| Grifola frondosa | Grifr_OBZ78918 | 72.3 | 0 | 1290 |
| Auricularia subglabra | Aurde3_1_99027 | 74.6 | 0 | 1269 |
| Lentinula edodes B17 | Lened_B_1_1_16258 | 70.6 | 0 | 1267 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_357091.T0 |
| Description | Chitin synthase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd04190 | Chitin_synth_C | - | 172 | 492 |
| Pfam | PF08407 | Chitin synthase N-terminal | IPR013616 | 107 | 175 |
| Pfam | PF01644 | Chitin synthase | IPR004834 | 176 | 335 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 485 | 504 | 19 |
| 2 | 525 | 547 | 22 |
| 3 | 567 | 589 | 22 |
| 4 | 601 | 623 | 22 |
| 5 | 648 | 670 | 22 |
| 6 | 779 | 801 | 22 |
| 7 | 821 | 843 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR013616 | Chitin synthase N-terminal |
| IPR029044 | Nucleotide-diphospho-sugar transferases |
| IPR004835 | Chitin synthase |
| IPR004834 | Fungal chitin synthase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004100 | chitin synthase activity | MF |
| GO:0016758 | transferase activity, transferring hexosyl groups | MF |
| GO:0006031 | chitin biosynthetic process | BP |
KEGG
| KEGG Orthology |
|---|
| K00698 |
EggNOG
| COG category | Description |
|---|---|
| M | Chitin synthase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GT | GT2 | GT2_Chitin_synth_1 |
Transcription factor
| Group |
|---|
| No records |
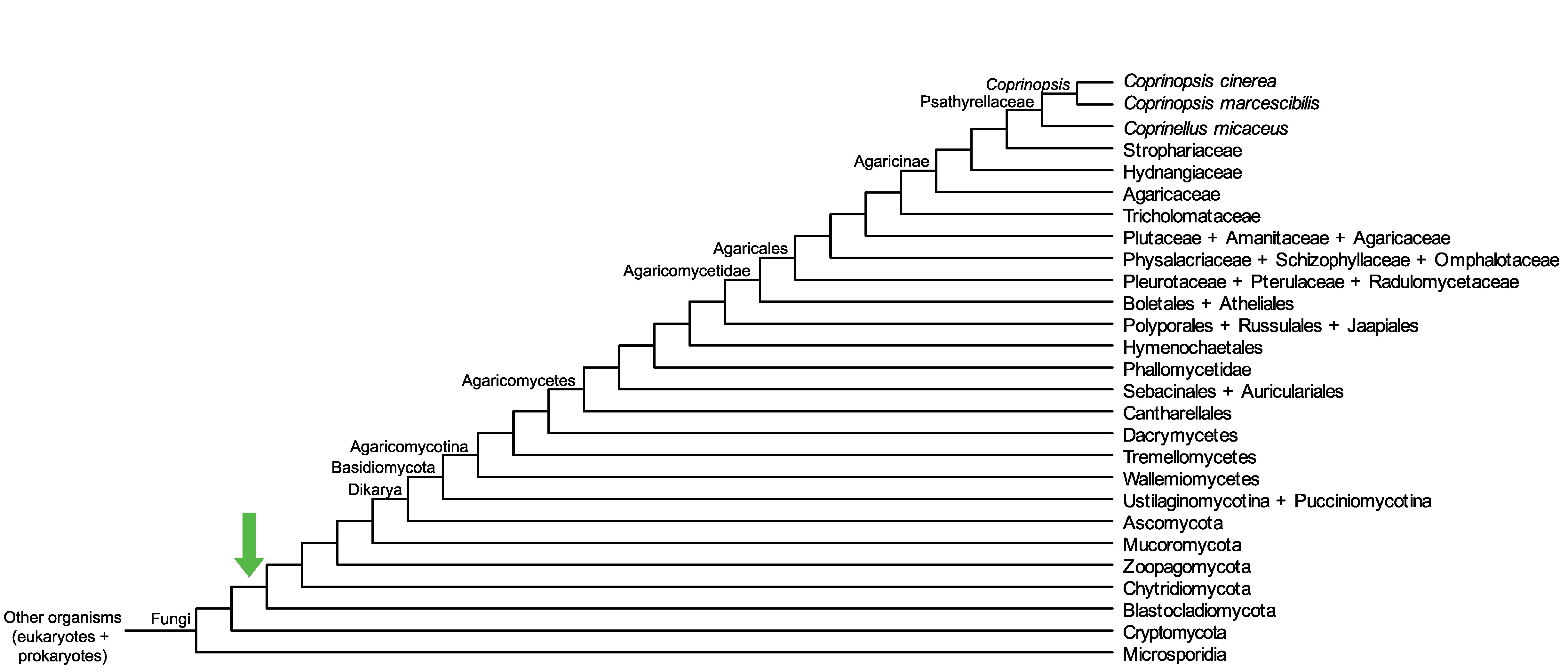
Conservation of CopciAB_357091 across fungi.
Arrow shows the origin of gene family containing CopciAB_357091.
Protein
| Sequence id | CopciAB_357091.T0 |
|---|---|
| Sequence |
>CopciAB_357091.T0 MAYRPSHNDPYYNQPSYPPATGYGDSSNWDAKSTKSYQSSYAGSQAHLNPSSYEMQQVNQHPVPAMPYNGGYPPQ QTMPSPGGFRDNGSVAGWSLARDKLMKRRSIKKVFLTQGNLVLDVPVPSHIVPVGKQDLEEFSKMRYTAATCDPD DFKASRYTLRPYLMGRETELFIVMTMYNEDEVLFVRTMNAVLKNVAHLCSRQKSKTWGPEGWKKVVVCVVSDGRN KVNKRTLQVLNLMGCYQEGIAKDSVAGKDVTAHIFEYTTTAIVTETGEVSQNTCPVQVLFCLKEQNKKKLNSHRW FFNAFGPLLNPNVCILLDVGTKPTGTSIYELWKCFDKHKHVGGACGEITVDSGRGCSLLLTSPLAASQNFEYKMS NILDKPLESVFGYISVLPGAFSAYRYKALLNGPDGKGPLASYFKGEAMHGGGSDAGLFERNMYLAEDRILCFEIV TKKKEAWTLKYVKSAKASTDVPATVPEFISQRRRWLNGSLFASIHATVFFFRIWTSGQGFFRKIALQFEFIYNAV QLLFTWTSLANFYLAFFFLVSSATAEGSTDAFNFLSTGAGKIVFQIFLNLYIGLLFVVLVCSLGNRPQGSKWTYT IAMFLFGLCNIITTWCAAYTVYLAVPHDIEGWRNFPELFKENRTLQEIVVAVLATYGMYFISSFIHFEPWHMFTS FAQYMFLLPSYVNILMMYAMCNLHDVTWGTKGDNGAAKDLGGAKKVKGEDGKEMMEVAVPTAREDVDQLWAASRA ALKVKPPEQKESRDAATKQADHDRNSRTNVVLAWVGSNMVLIIVFTSQAFLDWVNQNTSLGGSAFNPYLQFLFFA FAGLSAIRFTGSVVYLIFRLFGH |
| Length | 848 |
Coding
| Sequence id | CopciAB_357091.T0 |
|---|---|
| Sequence |
>CopciAB_357091.T0 ATGGCTTATCGTCCATCCCACAACGATCCTTACTACAATCAACCGAGCTATCCTCCAGCTACCGGTTACGGAGAT TCCTCAAATTGGGACGCAAAGTCCACAAAGTCGTACCAAAGTAGTTATGCAGGTTCCCAAGCCCACTTGAATCCA TCCAGCTACGAGATGCAACAGGTTAACCAGCACCCAGTTCCTGCAATGCCATACAACGGCGGCTACCCACCACAG CAAACTATGCCTTCCCCAGGCGGCTTTAGGGACAATGGCTCTGTCGCGGGCTGGTCCCTGGCCAGAGACAAGCTT ATGAAAAGGCGGTCTATCAAGAAAGTCTTCCTCACACAGGGAAACCTCGTCCTTGATGTACCCGTTCCCTCCCAC ATCGTCCCTGTTGGCAAGCAGGATCTCGAAGAATTCTCCAAGATGCGCTACACGGCGGCGACCTGCGACCCTGAC GACTTCAAAGCTTCTCGCTACACCCTCAGGCCCTACCTCATGGGCCGCGAGACCGAGCTCTTTATTGTCATGACC ATGTACAACGAAGACGAAGTCCTATTCGTCCGCACTATGAATGCCGTCCTCAAGAACGTTGCTCATTTATGCAGC CGTCAAAAGTCAAAAACTTGGGGTCCAGAAGGCTGGAAGAAGGTCGTCGTCTGTGTCGTGTCAGATGGTCGTAAC AAGGTCAACAAGCGGACACTGCAAGTATTGAACTTGATGGGATGCTACCAAGAAGGTATCGCTAAAGATTCCGTG GCGGGCAAGGATGTCACAGCCCACATCTTCGAGTACACCACCACAGCTATCGTGACAGAGACCGGCGAAGTCAGC CAGAACACATGTCCAGTCCAAGTTCTCTTCTGCTTGAAGGAACAGAACAAGAAGAAGTTGAACAGCCATAGATGG TTCTTCAATGCATTCGGACCTTTGCTGAACCCTAATGTCTGTATCCTCCTGGACGTCGGCACAAAGCCCACCGGA ACTTCAATCTATGAGCTATGGAAATGCTTCGATAAGCACAAACATGTCGGTGGTGCTTGTGGCGAAATCACTGTT GACAGTGGCCGCGGGTGCAGCCTCCTTCTCACAAGTCCTCTCGCAGCCTCCCAGAACTTTGAGTACAAAATGTCC AATATCCTTGACAAACCTTTGGAGAGCGTGTTTGGTTACATTAGCGTGTTGCCTGGTGCATTCAGCGCTTACCGA TACAAGGCTCTTTTGAACGGCCCTGATGGAAAGGGCCCGTTGGCATCTTACTTCAAGGGTGAGGCCATGCACGGA GGAGGTAGCGACGCTGGTCTCTTCGAGCGTAACATGTACCTCGCCGAAGATCGTATCCTCTGCTTCGAGATTGTC ACCAAGAAGAAGGAGGCCTGGACATTGAAGTATGTCAAGAGCGCAAAGGCATCTACTGACGTCCCCGCGACTGTC CCGGAATTCATCTCTCAACGTCGTCGATGGTTGAACGGTTCCTTATTCGCTTCCATCCATGCAACGGTTTTCTTC TTCAGGATCTGGACGTCTGGTCAGGGCTTCTTCCGCAAGATCGCTCTCCAATTCGAGTTCATCTACAATGCCGTC CAACTTCTCTTCACATGGACATCACTGGCCAACTTTTACCTCGCTTTCTTCTTCTTGGTTTCCTCGGCTACGGCA GAGGGCTCTACGGACGCATTCAACTTTCTGAGCACTGGTGCAGGCAAGATCGTGTTCCAAATCTTCCTCAATCTC TACATCGGTCTTCTGTTCGTCGTCTTGGTCTGTTCTCTGGGTAATCGACCTCAAGGTTCAAAATGGACGTACACA ATCGCCATGTTCTTGTTTGGTTTGTGTAATATCATCACCACATGGTGCGCTGCCTACACTGTCTACCTGGCTGTC CCCCACGATATCGAAGGATGGAGAAACTTCCCTGAACTGTTCAAGGAGAACCGCACGCTTCAGGAGATCGTCGTC GCCGTCCTCGCCACCTACGGCATGTACTTCATCAGTTCCTTCATCCACTTTGAACCTTGGCACATGTTCACCTCC TTTGCGCAATACATGTTCCTCCTCCCCTCGTACGTCAACATCCTCATGATGTACGCCATGTGCAACTTGCATGAT GTCACTTGGGGAACGAAAGGAGACAACGGCGCAGCGAAGGATTTGGGCGGCGCGAAGAAGGTGAAGGGAGAGGAT GGCAAGGAGATGATGGAAGTTGCGGTGCCTACTGCGAGGGAGGACGTGGATCAACTTTGGGCCGCGTCGAGGGCT GCGTTGAAGGTCAAGCCCCCGGAGCAGAAGGAGAGTCGAGATGCCGCTACCAAGCAGGCTGATCATGATCGGAAT AGTAGGACGAATGTTGTGTTGGCGTGGGTTGGTTCGAACATGGTGCTTATCATTGTTTTCACTTCCCAAGCCTTC TTGGACTGGGTGAACCAGAACACTTCGCTCGGTGGAAGTGCTTTCAACCCATACCTGCAATTCCTCTTCTTTGCC TTTGCCGGCCTATCAGCCATCCGCTTCACAGGTTCAGTCGTATACCTGATATTCAGACTATTCGGTCACTAG |
| Length | 2547 |
Transcript
| Sequence id | CopciAB_357091.T0 |
|---|---|
| Sequence |
>CopciAB_357091.T0 AAGCTTTCTTCCCAACGCCCACATCGACGGCCTCGTCGCCTCGTCCAGTCTTTCAAATTCCCTCACGAGCGTTGC CAACGCTATTCCATTCTTTTTCACAGCCGGTCTGGTCAACATATCTACCTAATATCCCTACGGACGCCAGCCACG ACTCACAAACACAATGGCTTATCGTCCATCCCACAACGATCCTTACTACAATCAACCGAGCTATCCTCCAGCTAC CGGTTACGGAGATTCCTCAAATTGGGACGCAAAGTCCACAAAGTCGTACCAAAGTAGTTATGCAGGTTCCCAAGC CCACTTGAATCCATCCAGCTACGAGATGCAACAGGTTAACCAGCACCCAGTTCCTGCAATGCCATACAACGGCGG CTACCCACCACAGCAAACTATGCCTTCCCCAGGCGGCTTTAGGGACAATGGCTCTGTCGCGGGCTGGTCCCTGGC CAGAGACAAGCTTATGAAAAGGCGGTCTATCAAGAAAGTCTTCCTCACACAGGGAAACCTCGTCCTTGATGTACC CGTTCCCTCCCACATCGTCCCTGTTGGCAAGCAGGATCTCGAAGAATTCTCCAAGATGCGCTACACGGCGGCGAC CTGCGACCCTGACGACTTCAAAGCTTCTCGCTACACCCTCAGGCCCTACCTCATGGGCCGCGAGACCGAGCTCTT TATTGTCATGACCATGTACAACGAAGACGAAGTCCTATTCGTCCGCACTATGAATGCCGTCCTCAAGAACGTTGC TCATTTATGCAGCCGTCAAAAGTCAAAAACTTGGGGTCCAGAAGGCTGGAAGAAGGTCGTCGTCTGTGTCGTGTC AGATGGTCGTAACAAGGTCAACAAGCGGACACTGCAAGTATTGAACTTGATGGGATGCTACCAAGAAGGTATCGC TAAAGATTCCGTGGCGGGCAAGGATGTCACAGCCCACATCTTCGAGTACACCACCACAGCTATCGTGACAGAGAC CGGCGAAGTCAGCCAGAACACATGTCCAGTCCAAGTTCTCTTCTGCTTGAAGGAACAGAACAAGAAGAAGTTGAA CAGCCATAGATGGTTCTTCAATGCATTCGGACCTTTGCTGAACCCTAATGTCTGTATCCTCCTGGACGTCGGCAC AAAGCCCACCGGAACTTCAATCTATGAGCTATGGAAATGCTTCGATAAGCACAAACATGTCGGTGGTGCTTGTGG CGAAATCACTGTTGACAGTGGCCGCGGGTGCAGCCTCCTTCTCACAAGTCCTCTCGCAGCCTCCCAGAACTTTGA GTACAAAATGTCCAATATCCTTGACAAACCTTTGGAGAGCGTGTTTGGTTACATTAGCGTGTTGCCTGGTGCATT CAGCGCTTACCGATACAAGGCTCTTTTGAACGGCCCTGATGGAAAGGGCCCGTTGGCATCTTACTTCAAGGGTGA GGCCATGCACGGAGGAGGTAGCGACGCTGGTCTCTTCGAGCGTAACATGTACCTCGCCGAAGATCGTATCCTCTG CTTCGAGATTGTCACCAAGAAGAAGGAGGCCTGGACATTGAAGTATGTCAAGAGCGCAAAGGCATCTACTGACGT CCCCGCGACTGTCCCGGAATTCATCTCTCAACGTCGTCGATGGTTGAACGGTTCCTTATTCGCTTCCATCCATGC AACGGTTTTCTTCTTCAGGATCTGGACGTCTGGTCAGGGCTTCTTCCGCAAGATCGCTCTCCAATTCGAGTTCAT CTACAATGCCGTCCAACTTCTCTTCACATGGACATCACTGGCCAACTTTTACCTCGCTTTCTTCTTCTTGGTTTC CTCGGCTACGGCAGAGGGCTCTACGGACGCATTCAACTTTCTGAGCACTGGTGCAGGCAAGATCGTGTTCCAAAT CTTCCTCAATCTCTACATCGGTCTTCTGTTCGTCGTCTTGGTCTGTTCTCTGGGTAATCGACCTCAAGGTTCAAA ATGGACGTACACAATCGCCATGTTCTTGTTTGGTTTGTGTAATATCATCACCACATGGTGCGCTGCCTACACTGT CTACCTGGCTGTCCCCCACGATATCGAAGGATGGAGAAACTTCCCTGAACTGTTCAAGGAGAACCGCACGCTTCA GGAGATCGTCGTCGCCGTCCTCGCCACCTACGGCATGTACTTCATCAGTTCCTTCATCCACTTTGAACCTTGGCA CATGTTCACCTCCTTTGCGCAATACATGTTCCTCCTCCCCTCGTACGTCAACATCCTCATGATGTACGCCATGTG CAACTTGCATGATGTCACTTGGGGAACGAAAGGAGACAACGGCGCAGCGAAGGATTTGGGCGGCGCGAAGAAGGT GAAGGGAGAGGATGGCAAGGAGATGATGGAAGTTGCGGTGCCTACTGCGAGGGAGGACGTGGATCAACTTTGGGC CGCGTCGAGGGCTGCGTTGAAGGTCAAGCCCCCGGAGCAGAAGGAGAGTCGAGATGCCGCTACCAAGCAGGCTGA TCATGATCGGAATAGTAGGACGAATGTTGTGTTGGCGTGGGTTGGTTCGAACATGGTGCTTATCATTGTTTTCAC TTCCCAAGCCTTCTTGGACTGGGTGAACCAGAACACTTCGCTCGGTGGAAGTGCTTTCAACCCATACCTGCAATT CCTCTTCTTTGCCTTTGCCGGCCTATCAGCCATCCGCTTCACAGGTTCAGTCGTATACCTGATATTCAGACTATT CGGTCACTAGATTTCAAGAATAATCTCTCCCCCCATCTTGCATACGCTACCATCTACCGCTACCCACGACACATA TAATCGCTCACCCATCCTCAGCTGGGTTCACTCTTTTACAAAAAGAAGGATCTACCGCTAATACCTCTAAATCAT CCGTTGTACTTTTACTACCCATCCTCTACGATCGATACCCTACATACCCCGCAGGTTGATCCTAGCTCATGTTGT AGGAAATTTCTTTCTATCTTTCTTGTTCTATTTGTTATTTTTCGTCTCGTCCCCCCTTTTTACATGATTGCATTT GTTTCTTTACGTTCTTTTTGTTGGCTGGTGGCTTTCCTTCGAGTCCATCGAAGGAGTCCACTGGTCGATACCTTT TTCGATACGAATTCGATTCGACGTTCCTTCTCGTTCGTTGCATCTGTTTCGTGATGTACTTGGACTTTGACGAAT CTGCAACTCCGTTTGCTCTCTC |
| Length | 3172 |
Gene
| Sequence id | CopciAB_357091.T0 |
|---|---|
| Sequence |
>CopciAB_357091.T0 AAGCTTTCTTCCCAACGCCCACATCGACGGCCTCGTCGCCTCGTCCAGTCTTTCAAATTCCCTCACGAGCGTTGC CAACGCTATTCCATTCTTTTTCACAGCCGGTCTGGTCAACATATCTACCTAATATCCCTACGGACGCCAGCCACG ACTCACAAACACAATGGCTTATCGTCCATCCCACAACGATCCTTACTACAATCAACCGAGCTATCCTCCAGCTAC CGGTTACGGAGATTCCTCAAATTGGGACGCAAAGTCCACAAAGTCGTACCAAAGTAGTTATGCAGGTTCCCAAGC CCACTTGAATCCATCCAGCTACGAGATGCAACAGGTTAACCAGCACCCAGTTCCTGCAATGCCATACAACGGCGG CTACCCACCACAGCAAACTATGCCTTCCCCAGGCGGCTTTAGGGACAATGGCTCTGTCGCGGGCTGGTCCCTGGC CAGAGACAAGCTTATGAAAAGGCGGTCTATCAAGAAAGTCTTCCTCACACAGGGAAACCTCGTCCTTGATGTACC CGTTCCCTCCCACATCGTCCCTGTTGGCAAGCAGGATCTCGAAGAATTCTCCAAGATGCGCTACACGGCGGCGAC CTGCGACCCTGACGACTTCAAAGCTTCTCGCTACACCCTCAGGCCCTACCTCATGGGCCGCGAGACCGAGCTCTT TATTGTCATGACCATGTACAACGAAGACGAAGTCCTATTCGTCCGCACTATGAATGCGTACGTGTTTATTGCCCT CTGTGAGATTCATAATGTATCTCTGACCCATTATACAGCGTCCTCAAGAACGTTGCTCATTTATGCAGCCGTCAA AAGTCAAAAACTTGGGGTCCAGAAGGCTGGAAGAAGGTCGTCGTCTGTGTCGTGTCAGATGGTCGTAACAAGGTC AACAAGCGGACACTGCAAGTATTGAACTTGGTAAGTTTCATCGTTCCCATCAGATATTATCCCGTCCTAACCACC CAACGTTGGCAGATGGGATGCTACCAAGAAGGTATCGCTAAAGATTCCGTGGCGGGCAAGGATGTCACAGCCCAC ATCTTCGAGTATGTTATCTCCATCTCCCGTCTTTCAGAACACGGGCTCAACTCCCCGTAGGTACACCACCACAGC TATCGTGACAGAGACCGGCGAAGTCAGCCAGAACACATGTCCAGTCCAAGTTCTCTTCTGCTTGAAGGAACAGAA CAAGAAGAAGTTGAACAGCCATAGATGGTTCTTCAATGCATTCGGACCTTTGCTGAACCCTAATGGTAAGTGCTA TATTTAGCGTACAACGATGGATATCCTTGTTAATCGCATGCCTATAGTCTGTATCCTCCTGGACGTCGGCACAAA GCCCACCGGAACTTCAATCTATGAGCTATGGAAATGTAAGGTCTATCTATATCGTCTGGGAATTTCAACTCAACT CGGCCCACCCAGGCTTCGATAAGCACAAACATGTCGGTGGTGCTTGTGGCGAGTAAGTAACCCCGCCGTATACTC ATCGGTCTTGTTCTGATCGTCTTCTCCAGAATCACTGTTGACAGTGGCCGCGGGTGCAGCCTCCTTCTCACAAGT CCTCTCGCAGCCTCCCAGAACTTTGAGGTATAACCACCCTTCCTCTTTCTAGAAGTCCTCGTCGTCAAGTGCGTT AACAACGTGTCCGTGTTTCCTGTTCACAGTACAAAATGTCCAATATCCTTGACAAACCTTTGGAGGTGGGTTGAC ATGGTTATTTCATTGAGGACCGTGGTTATTGATTCAAATTACGTGGTCTAGAGCGTGTTTGGTTACATTAGCGTG TTGCCTGGTGCATTCAGCGCTTACCGATACAAGGCTCTTTTGAACGGCCCTGATGGAAAGGGCCCGTTGGCATCT TACTTCAAGGGTGAGGCCATGCACGGAGGAGGTAGCGACGCTGGTCTCTTCGGTAAGTGAACATCCTAGAAATCC GATGCGATGGACGTTGGCTGACTCTCTTTTCAGAGGTAATGCCATTATTATCGAGTCGGAGGGATGACGCTGAAT GTTCCGTTTCCAGCGTAACATGTACCTCGCCGAAGATCGTATCCTCTGCTTCGAGATTGTCACCAAGAAGAAGGA GGCCTGGACGTAAGTCTCCCACTTCATCTGTATCCTATCCACTTGAAGCTTATTGTGTTCTATTAATAGATTGAA GTATGTCAAGAGCGCAAAGGCATCTACTGACGTCCCCGCGACTGTCCCGGAATTCATCTCTCAACGTCGTCGATG GTTGAACGGTTCCTTATTCGCTTCCATCCATGCAACGGTTTTCTTCTTCAGGATCTGGACGTCTGGTCAGGGCTT CTTCCGCAAGATCGCTCTCCAATTCGAGTTCATCTACAATGCCGTCCAACTTCTCTTCACATGGACATCACTGGC CAACTTTTACCTCGCTTTCTTCTTCGTAAGCTGGTTTTTAGTTCCAACACATGCCAGTCGCTGATCGAGCATTGA TTGTAGTTGGTTTCCTCGGCTACGGCAGAGGGCTCTACGGACGCATTCAACTTTCTGAGCACTGGTGCAGGCAAG ATCGTGTTCCAAATCTTCCTCAATCTCTACATCGGTCTTCTGTTCGTCGTCTTGGTCTGTTCTCTGGGTAATCGA CCTCAAGTATGTACCATCCGCATAATGCGTGTTTCTGGCGATCGTCTTACTGATTTGAATCGTTTAGGGTTCAAA ATGGACGTACACAATCGCCATGTTCTTGTTTGGTTTGTGTAATATCATCACCACATGGTGCGCTGCCTACACTGT CTACCTGGCTGTCCCCCACGATATCGAAGGATGGAGAAACTTCCCTGAGTAAGTCTACTTCCTGTTTCAAGATGC AAGACGTCATTGATTCCTTTTCCACAGACTGTTCAAGGAGAACCGCACGCTTCAGGAGATCGTCGTCGCCGTCCT CGCCACCTACGGCATGTACTTCATCAGTTCCTTCATCCACTTTGAACCTTGGCACATGTTCACCTCCTTTGCGCA ATACATGTTCCTCCTCCCCTCGTACGTCAACATCCTCATGATGTACGCCATGTGCAACTTGCATGATGTCACTTG GGGAACGAAAGGAGACAACGGCGCAGCGAAGGATTTGGGCGGCGCGAAGAAGGTGAAGGGAGAGGATGGCAAGGA GATGATGGAAGTTGCGGTGCCTACTGCGAGGGAGGACGTGGATCAACTTTGGGCCGCGTCGAGGGCTGCGTTGAA GGTCAAGCCCCCGGAGCAGAAGGAGAGTCGAGATGCCGCTACCAAGCAGGCTGATCATGATCGGAATAGTAGGAC GAATGTTGTGTTGGCGTGGGTTGGTTCGAACATGTGAGTGCAGTCGACGGTTCTAGATGCTTCTCGTTGGCGGAT GACTGACTGATCTTTTTGCAGGGTGCTTATCATTGTTTTCACTTCCCAAGCCTTCTTGGACTGGGTGAACCAGAA CACTTCGCTCGGTGGAAGTGCTTTCAACCCATACCTGCAATTCCTCTTCTTTGCCTTGTACGTCCTCCTCCTTTG ACAACCCTATGCCCAGGCTCAGACACCTTTCTTTCTCTAGTGCCGGCCTATCAGCCATCCGCTTCACAGGTTCAG TCGTATACCTGATATTCAGACTATTCGGTCACTAGATTTCAAGAATAATCTCTCCCCCCATCTTGCATACGCTAC CATCTACCGCTACCCACGACACATATAATCGCTCACCCATCCTCAGCTGGGTTCACTCTTTTACAAAAAGAAGGA TCTACCGCTAATACCTCTAAATCATCCGTTGTACTTTTACTACCCATCCTCTACGATCGATACCCTACATACCCC GCAGGTTGATCCTAGCTCATGTTGTAGGAAATTTCTTTCTATCTTTCTTGTTCTATTTGTTATTTTTCGTCTCGT CCCCCCTTTTTACATGATTGCATTTGTTTCTTTACGTTCTTTTTGTTGGCTGGTGGCTTTCCTTCGAGTCCATCG AAGGAGTCCACTGGTCGATACCTTTTTCGATACGAATTCGATTCGACGTTCCTTCTCGTTCGTTGCATCTGTTTC GTGATGTACTTGGACTTTGACGAATCTGCAACTCCGTTTGCTCTCTC |
| Length | 4097 |