CopciAB_359952
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_359952 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 359952 |
| Uniprot id | Functional description | Lytic transglycolase | |
| Location | scaffold_1:2561139..2562472 | Strand | - |
| Gene length (nt) | 1334 | Transcript length (nt) | 1169 |
| CDS length (nt) | 759 | Protein length (aa) | 252 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | NCU02197 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7266516 | 55.9 | 2.849E-80 | 265 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB29499 | 46.2 | 4.97E-75 | 250 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1096037 | 42.9 | 8.521E-65 | 220 |
| Lentinula edodes NBRC 111202 | Lenedo1_1165591 | 41.8 | 4.882E-64 | 218 |
| Lentinula edodes B17 | Lened_B_1_1_3462 | 42.3 | 8.558E-64 | 217 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1502629 | 42.1 | 1.09E-63 | 217 |
| Grifola frondosa | Grifr_OBZ80003 | 40.2 | 5.745E-61 | 209 |
| Auricularia subglabra | Aurde3_1_1271686 | 44.8 | 6.356E-57 | 198 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_148241 | 40.2 | 7.974E-57 | 197 |
| Pleurotus ostreatus PC9 | PleosPC9_1_54071 | 40.8 | 7.72E-46 | 165 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_140389 | 38.2 | 4.111E-42 | 154 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_1904 | 62.6 | 1.488E-38 | 144 |
| Schizophyllum commune H4-8 | Schco3_38802 | 53.8 | 2.867E-29 | 117 |
| Flammulina velutipes | Flave_chr08AA00894 | 27.4 | 3.227E-16 | 78 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_359952.T0 |
| Description | Lytic transglycolase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd22191 | DPBB_RlpA_EXP_N-like | - | 150 | 249 |
| Pfam | PF03330 | Lytic transglycolase | IPR009009 | 148 | 245 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.7735 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 5 | 27 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR036908 | RlpA-like domain superfamily |
| IPR009009 | RlpA-like protein, double-psi beta-barrel domain |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | Lytic transglycolase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| EXPN | EXPN |
Transcription factor
| Group |
|---|
| No records |
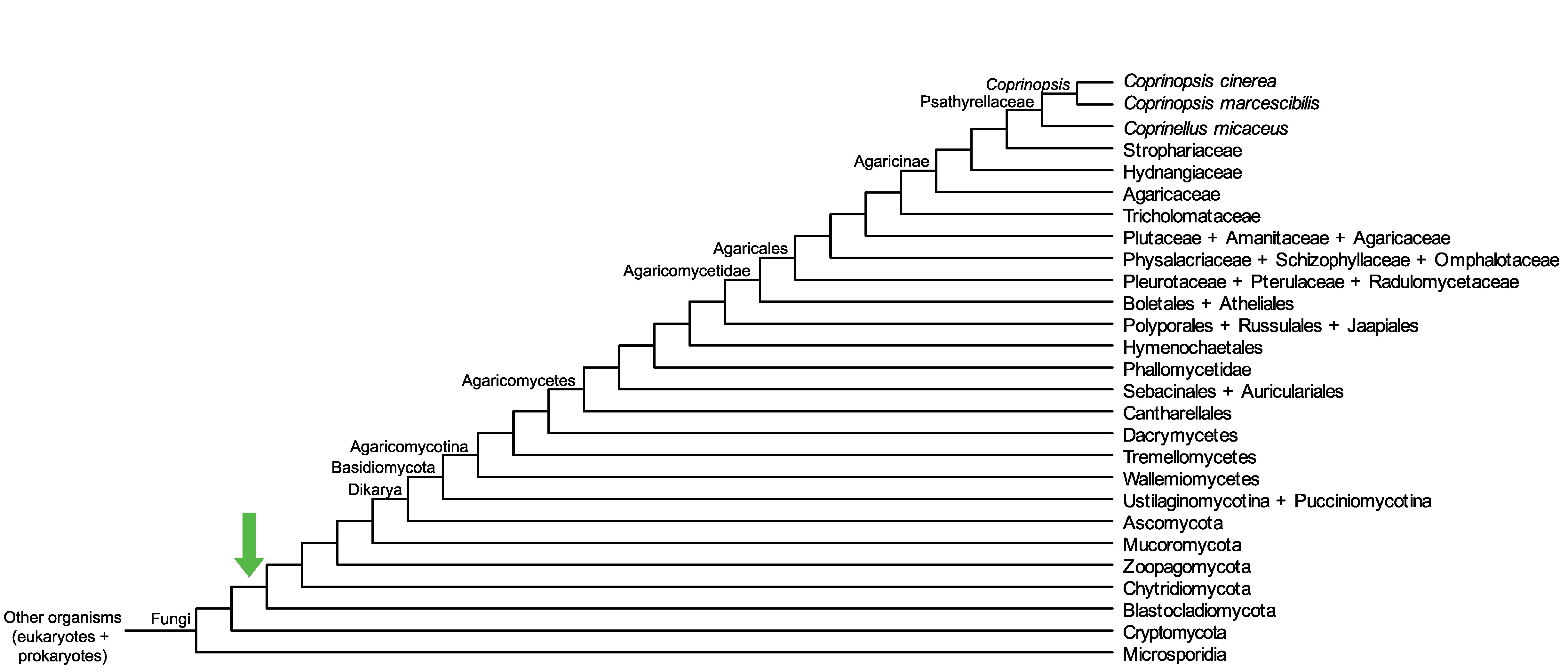
Conservation of CopciAB_359952 across fungi.
Arrow shows the origin of gene family containing CopciAB_359952.
Protein
| Sequence id | CopciAB_359952.T0 |
|---|---|
| Sequence |
>CopciAB_359952.T0 MPSFVNAVVVFLAAVASVNGLVTPHIARHNAHHRSIAARVAEPQVQPALVPRAPTATLRKRCRPRLVFVTPSPSP STAPAPSPEVPPVNVAPSPSPEPQPEPEQPAPVPSPEAPPAPVEPAPQPAPQQPAPQPGNGGGNQPGFMSGTQTG QGTFYDAGLGACGIVNTNADYIAAVSHLLFDNYPGYNRVNPNTNPLCGRTVTATYQGRSVTVAITDRCEGCAVTD LDFTPSAFAQLADHSLGRISGVTWHWN |
| Length | 252 |
Coding
| Sequence id | CopciAB_359952.T0 |
|---|---|
| Sequence |
>CopciAB_359952.T0 ATGCCTTCCTTTGTCAACGCTGTCGTCGTTTTCCTGGCCGCCGTCGCCTCCGTCAACGGCCTCGTCACTCCCCAC ATCGCCAGGCACAACGCGCACCACAGGTCAATCGCTGCCCGCGTCGCTGAGCCTCAGGTCCAGCCCGCGCTCGTC CCCAGGGCTCCAACAGCCACTCTCAGGAAGCGATGCAGGCCCAGGCTCGTATTCGTTACCCCCTCTCCTTCCCCC AGCACCGCCCCAGCTCCCAGCCCTGAGGTCCCTCCTGTCAACGTTGCACCCTCCCCGTCCCCAGAGCCCCAGCCT GAGCCTGAGCAGCCCGCACCTGTCCCGTCCCCGGAGGCCCCTCCTGCACCTGTCGAACCTGCTCCCCAACCTGCC CCACAGCAACCTGCCCCACAGCCCGGCAACGGCGGCGGTAACCAGCCAGGGTTCATGTCTGGCACACAGACCGGC CAAGGCACCTTCTACGACGCTGGTCTTGGTGCCTGCGGTATTGTGAACACCAACGCCGACTACATTGCTGCCGTC TCCCACCTCCTCTTCGATAACTACCCCGGATACAACCGTGTCAACCCTAACACCAACCCCCTCTGCGGCAGGACT GTCACCGCTACCTACCAAGGAAGGAGCGTCACCGTCGCCATCACTGACCGCTGCGAGGGCTGTGCAGTCACTGAC CTTGACTTCACCCCTTCCGCGTTCGCCCAACTTGCCGACCACAGCCTCGGCCGCATCAGCGGTGTTACCTGGCAC TGGAACTAG |
| Length | 759 |
Transcript
| Sequence id | CopciAB_359952.T0 |
|---|---|
| Sequence |
>CopciAB_359952.T0 ACCGCTCGTTCTTCAGCCATGCCTTCCTTTGTCAACGCTGTCGTCGTTTTCCTGGCCGCCGTCGCCTCCGTCAAC GGCCTCGTCACTCCCCACATCGCCAGGCACAACGCGCACCACAGGTCAATCGCTGCCCGCGTCGCTGAGCCTCAG GTCCAGCCCGCGCTCGTCCCCAGGGCTCCAACAGCCACTCTCAGGAAGCGATGCAGGCCCAGGCTCGTATTCGTT ACCCCCTCTCCTTCCCCCAGCACCGCCCCAGCTCCCAGCCCTGAGGTCCCTCCTGTCAACGTTGCACCCTCCCCG TCCCCAGAGCCCCAGCCTGAGCCTGAGCAGCCCGCACCTGTCCCGTCCCCGGAGGCCCCTCCTGCACCTGTCGAA CCTGCTCCCCAACCTGCCCCACAGCAACCTGCCCCACAGCCCGGCAACGGCGGCGGTAACCAGCCAGGGTTCATG TCTGGCACACAGACCGGCCAAGGCACCTTCTACGACGCTGGTCTTGGTGCCTGCGGTATTGTGAACACCAACGCC GACTACATTGCTGCCGTCTCCCACCTCCTCTTCGATAACTACCCCGGATACAACCGTGTCAACCCTAACACCAAC CCCCTCTGCGGCAGGACTGTCACCGCTACCTACCAAGGAAGGAGCGTCACCGTCGCCATCACTGACCGCTGCGAG GGCTGTGCAGTCACTGACCTTGACTTCACCCCTTCCGCGTTCGCCCAACTTGCCGACCACAGCCTCGGCCGCATC AGCGGTGTTACCTGGCACTGGAACTAGGTTTCCAACCTCGCAATAACCAACCCTCATAGAAACTCGTAACGAGTT CAAGCATACCCAAGCTTTCTCCTCTCTCTATCATTTCCTTTCCAAGAAAACGCTACTGCCAGCTCAGTGGCGCTA TTGCAATCATCAGGCTTTCTGGCACAAGTCGACACACACTTGCAAAGGGCGCTCTCCGTGCGAGCGATCGTTCAC CAGTTGGCTGCTTGCAGCTAACCTGACGTTCGAACGACTGGCCTTGTCATAGTTTAGTTTGCACTCGCTTTCGAG CCAGACTCGTTAGGATCATCACTGTCAGTCTTTTGTAACCTCAGTCATGTACTTGCTGTAGCCATGTATCATGCT TTCACTCTCTTGTTCTCGTCCTGGAATAAACTCATATGTTGTAT |
| Length | 1169 |
Gene
| Sequence id | CopciAB_359952.T0 |
|---|---|
| Sequence |
>CopciAB_359952.T0 ACCGCTCGTTCTTCAGCCATGCCTTCCTTTGTCAACGCTGTCGTCGTTTTCCTGGCCGCCGTCGCCTCCGTCAAC GGCCTCGTCACTCCCCACATCGCCAGGCACAACGCGCACCACAGGTCAATCGCTGCCCGCGTCGCTGAGCCTCAG GTCCAGCCCGCGCTCGTCCCCAGGGCTCCAACAGCCACTCTCAGGAAGCGATGCAGGCCCAGGCTCGTATTCGTT ACCCCCTCTCCTTCCCCCAGCACCGCCCCAGCTCCCAGCCCTGAGGTCCCTCCTGTCAACGTTGCACCCTCCCCG TCCCCAGAGCCCCAGCCTGAGCCTGAGCAGCCCGCACCTGTCCCGTCCCCGGAGGCCCCTCCTGCACCTGTCGAA CCTGCTCCCCAACCTGCCCCACAGCAACCTGCCCCACAGCCCGGCAACGGCGGCGGTAACCAGCCAGGGTTCATG TCTGGCACACAGACCGGCCAAGGCACCTTCTACGACGGTATGCAGCCTGATCTACTTGAGATGCACTTTTTCTAA CCTTTTTTCTTCGTCCCAGCTGGTCTTGGTGCCTGCGGTATTGTGAACACCAACGCCGACTACATTGCTGCCGTC TCCCACCTCCTCTTCGATAACTACCCGTGCGTATTATCTTTTAATGAATGAACTAGTCCTGATTACCATTGATCT TCCAGCGGATACAACCGTGTCAACCCTAACACCAACCCCCTCTGCGGCAGGACTGTCACCGCTACCTGTACGTCC ATGTTATTTCCTTGTCCATCGAATTGCTGTGTTCAACGACTTCTAGACCAAGGAAGGAGCGTCACCGTCGCCATC ACTGACCGCTGCGAGGGCTGTGCAGTCACTGACCTTGACTTCACCCCTTCCGCGTTCGCCCAACTTGCCGACCAC AGCCTCGGCCGCATCAGCGGTGTTACCTGGCACTGGAACTAGGTTTCCAACCTCGCAATAACCAACCCTCATAGA AACTCGTAACGAGTTCAAGCATACCCAAGCTTTCTCCTCTCTCTATCATTTCCTTTCCAAGAAAACGCTACTGCC AGCTCAGTGGCGCTATTGCAATCATCAGGCTTTCTGGCACAAGTCGACACACACTTGCAAAGGGCGCTCTCCGTG CGAGCGATCGTTCACCAGTTGGCTGCTTGCAGCTAACCTGACGTTCGAACGACTGGCCTTGTCATAGTTTAGTTT GCACTCGCTTTCGAGCCAGACTCGTTAGGATCATCACTGTCAGTCTTTTGTAACCTCAGTCATGTACTTGCTGTA GCCATGTATCATGCTTTCACTCTCTTGTTCTCGTCCTGGAATAAACTCATATGTTGTAT |
| Length | 1334 |