CopciAB_364173
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_364173 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | IRE1 | Synonyms | 364173 |
| Uniprot id | Functional description | domain in protein kinases, N-glycanases and other nuclear proteins | |
| Location | scaffold_9:1316832..1320960 | Strand | - |
| Gene length (nt) | 4129 | Transcript length (nt) | 3843 |
| CDS length (nt) | 3477 | Protein length (aa) | 1158 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN0235_ireA |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB19766 | 57.7 | 0 | 1284 |
| Agrocybe aegerita | Agrae_CAA7269916 | 57 | 0 | 1238 |
| Schizophyllum commune H4-8 | Schco3_2629108 | 55.7 | 0 | 1234 |
| Flammulina velutipes | Flave_chr11AA01750 | 56.4 | 0 | 1174 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_37330 | 53.9 | 0 | 1158 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_120939 | 51.8 | 0 | 1114 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1060565 | 52.9 | 0 | 1105 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1471524 | 49.5 | 0 | 1095 |
| Lentinula edodes NBRC 111202 | Lenedo1_739048 | 48.9 | 0 | 1033 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_10463 | 48.9 | 0 | 1033 |
| Grifola frondosa | Grifr_OBZ76674 | 49.2 | 2.094E-304 | 1004 |
| Auricularia subglabra | Aurde3_1_1272235 | 48.1 | 4.079E-295 | 934 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_60620 | 74 | 7.946E-234 | 754 |
| Lentinula edodes B17 | Lened_B_1_1_11946 | 51.3 | 1.244E-225 | 730 |
| Pleurotus ostreatus PC9 | PleosPC9_1_82636 | 79.6 | 1.387E-224 | 727 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | IRE1 |
|---|---|
| Protein id | CopciAB_364173.T0 |
| Description | domain in protein kinases, N-glycanases and other nuclear proteins |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd09769 | Luminal_IRE1 | - | 63 | 384 |
| CDD | cd10422 | RNase_Ire1 | - | 1024 | 1153 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 714 | 906 |
| Pfam | PF06479 | Ribonuclease 2-5A | IPR010513 | 1026 | 1153 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.9802 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR018391 | Pyrrolo-quinoline quinone beta-propeller repeat |
| IPR045133 | Serine/threonine-protein kinase/endoribonuclease IRE1/2-like |
| IPR000719 | Protein kinase domain |
| IPR010513 | KEN domain |
| IPR015943 | WD40/YVTN repeat-like-containing domain superfamily |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR038357 | KEN domain superfamily |
| IPR011047 | Quinoprotein alcohol dehydrogenase-like superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004521 | endoribonuclease activity | MF |
| GO:0004674 | protein serine/threonine kinase activity | MF |
| GO:0030968 | endoplasmic reticulum unfolded protein response | BP |
| GO:0005524 | ATP binding | MF |
| GO:0004540 | ribonuclease activity | MF |
| GO:0006397 | mRNA processing | BP |
| GO:0005515 | protein binding | MF |
KEGG
| KEGG Orthology |
|---|
| K08852 |
EggNOG
| COG category | Description |
|---|---|
| T | domain in protein kinases, N-glycanases and other nuclear proteins |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
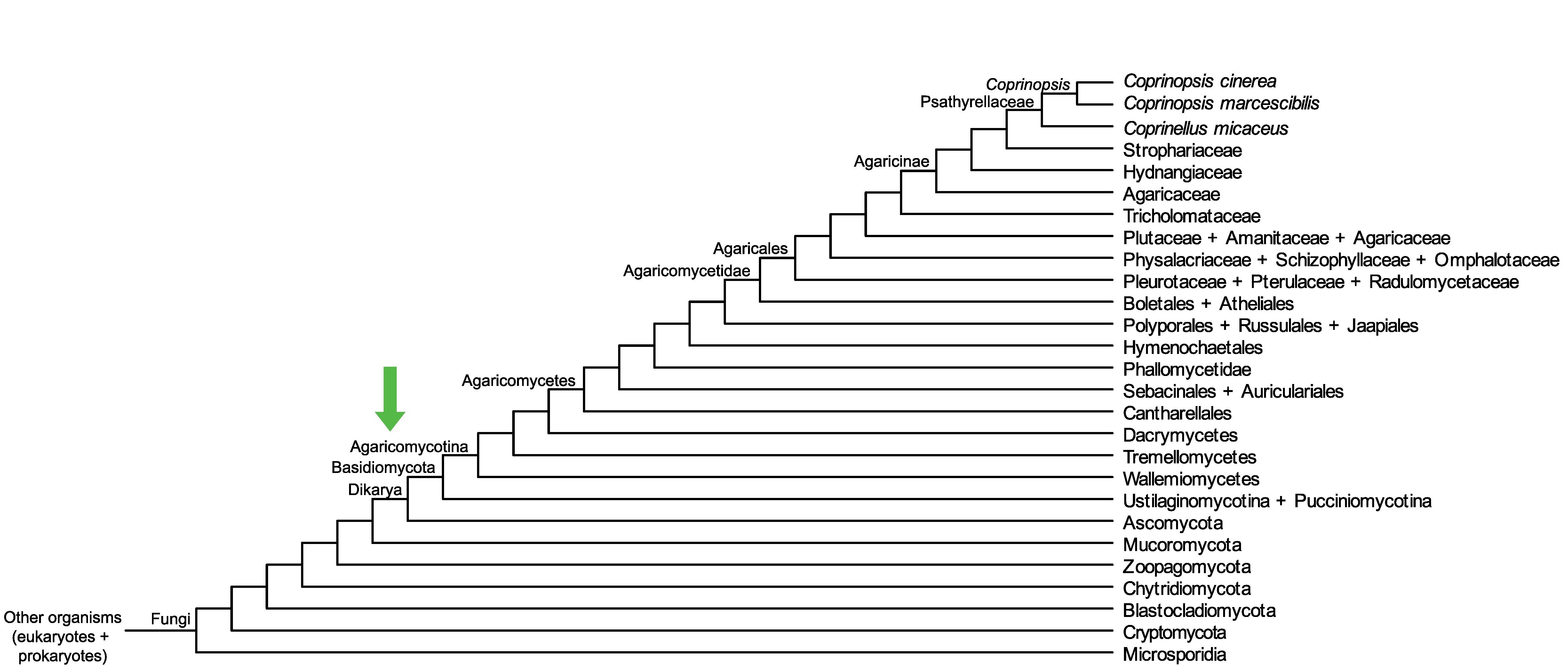
Conservation of CopciAB_364173 across fungi.
Arrow shows the origin of gene family containing CopciAB_364173.
Protein
| Sequence id | CopciAB_364173.T0 |
|---|---|
| Sequence |
>CopciAB_364173.T0 MAFVFFLQFLLLIVGAVCWAEIVNVRSTIPAAETAALIQRTRPAPTYATGGILEPTKDLELLDIVLVASIDGKFH ALSRSTGKILWSMPAFARAEGSSTLSSLAPLVRTTHPENDPDDFDDDTTPETYIIEPQSGHIYVVNSEDDSLQRF PFSMAELVDMSPFTSPGDDEKRVFIGRKETSLLLLELETGRIKATLNSECPPYDDRRIDLDELEEPESRGSSTTE VFIGRTDYYISIYPHRSKGYGTRAPVQNLSFSVYGPNNEDNLLQANYRRTSDDAYIQGFPNGDVYSFKTKSEGGR VQTQPLWVQKFHAPTVAVFDVLRGSTPLQPHPHAFVLLQPRPRAEDVIPGLDRSLLRGAKSAYVGMVEETGSLYA MSPSHYPLVAFGDAEEGRKGRLIEGSGGSAQSIHDIIRNQKEADRRIRCEDNPRDRECLLGMHPLDETDGHESRM RRLLEGPSSISPIPTFRPGQNQWIDHGIEVVPNIEAPPNNSQQPATPVPPEEVRKTGWEYLGVTLGLGLVSLWFL FKKFGQREGFKVPKFIPVPNGDAQIARPLAPIPDTATEPAPAPEPATVQPLQIIPEQPAPPAPLPLPLPPLEDPT TDALAGTGEEGDDTDREGDPEPETVATPGRKKPRRGKRGKKKKKDATPIPGEEGVEGKAPNGPDGPAANGTPNIN PPDNNPDATIAPAAPQSPSLVVSSPIIPAAAGPSLVVSDTILGFGSHGTVVFQGSLQGRAVAVKRLLKDFVTLAS REVSILQESDDHPNVIRYYYQEAHANFLYIALELCPASLADIIETPDREAFRDIAISFDPKRALKQITSGLKHLH ALKLVHRDIKPQNILITTNKSTGRGRPTYRMLISDFGLCKKLDVDQTSFLPTMNGGMGAGTVGWRAPEILRGEVK LDDLSDDHSMSSRGSVSTINGSSSSSSTSGLSTAKPTRLTKSVDIFALGCLYYYTLTNGSHPYGDRFEREVNILK DAKSLDLLERFGEEGTEACDLIEKMLDPEASERPDTTACLLHPFFWDPARRLNFLQDASDRFEIMCRDPKDPLLL QLETGALAIVGNDWHARLDRLFVENLGKFRKYDGKSVQDLLRALRNKKHHYQDLPDNVKRSLGPMPEGFLAYFTK RFPRLFLHVHRVISDTVLRTESMFRSYFELPDS |
| Length | 1158 |
Coding
| Sequence id | CopciAB_364173.T0 |
|---|---|
| Sequence |
>CopciAB_364173.T0 ATGGCGTTCGTATTCTTCCTACAGTTCTTGCTCCTCATCGTGGGCGCGGTCTGCTGGGCAGAGATCGTCAATGTC CGGTCAACCATCCCAGCAGCGGAGACGGCGGCGCTCATACAACGAACGAGACCAGCACCTACTTATGCGACCGGA GGGATTCTAGAACCCACCAAAGACCTAGAGCTGCTGGACATTGTCCTGGTAGCCTCGATCGACGGCAAATTCCAT GCACTCAGTCGCTCGACGGGCAAGATCTTGTGGTCTATGCCTGCATTCGCACGGGCAGAAGGCTCGTCAACTCTT TCAAGCTTGGCGCCTCTGGTTAGGACGACACACCCAGAAAACGACCCCGACGACTTTGACGACGATACAACACCA GAAACCTATATCATTGAACCGCAGTCTGGCCACATCTATGTCGTAAACTCAGAAGACGACTCGTTGCAACGGTTC CCGTTCTCAATGGCGGAGTTGGTTGACATGTCGCCGTTCACCTCCCCAGGTGACGACGAGAAGAGGGTATTCATT GGGCGCAAGGAGACTTCACTATTGTTACTGGAGCTCGAAACAGGAAGAATCAAGGCGACCCTCAACTCCGAATGT CCGCCTTATGACGACAGGCGTATTGACCTCGACGAGCTGGAAGAACCAGAGTCGCGGGGCTCATCGACCACCGAA GTATTCATTGGGCGAACAGATTATTACATTTCCATCTATCCCCACCGCTCTAAGGGCTACGGCACTCGGGCGCCC GTCCAAAACCTGTCATTCTCAGTCTACGGACCCAACAACGAAGACAACTTGCTCCAAGCAAACTATCGAAGGACT AGCGACGATGCGTACATCCAAGGTTTTCCCAACGGGGATGTCTACTCCTTTAAGACCAAGTCGGAAGGGGGTAGA GTCCAAACACAACCTTTATGGGTTCAGAAATTCCATGCACCGACGGTCGCTGTTTTTGATGTCCTACGGGGCTCA ACTCCACTTCAGCCACATCCTCACGCCTTTGTTCTCCTGCAACCACGACCACGCGCGGAAGATGTCATCCCAGGT CTCGACCGCTCTCTCTTACGTGGTGCAAAGTCGGCCTACGTTGGGATGGTGGAGGAAACGGGGAGCCTTTACGCT ATGAGCCCTTCACATTATCCACTGGTTGCGTTTGGGGACGCAGAGGAGGGCAGAAAGGGACGACTTATTGAGGGT TCTGGGGGTTCAGCGCAAAGCATCCATGATATCATCCGCAACCAGAAGGAGGCTGACAGGCGGATTAGATGCGAG GATAACCCGAGAGACCGCGAGTGTCTACTGGGTATGCATCCGCTTGACGAAACGGACGGCCATGAAAGTCGAATG CGACGACTGCTCGAAGGACCTTCAAGCATCAGCCCCATCCCGACGTTCAGACCCGGTCAGAACCAATGGATCGAT CACGGAATCGAAGTCGTCCCCAACATTGAAGCTCCACCCAACAATTCTCAACAACCTGCCACGCCTGTCCCCCCG GAGGAAGTACGGAAGACGGGATGGGAATATCTGGGAGTAACACTCGGGCTGGGTTTGGTGTCTTTGTGGTTCTTA TTCAAGAAGTTCGGCCAGCGAGAGGGATTCAAGGTACCCAAATTCATTCCCGTACCCAACGGGGACGCGCAGATC GCCCGTCCCCTTGCTCCCATACCGGACACTGCCACCGAGCCAGCACCAGCACCCGAACCAGCTACGGTCCAGCCT CTTCAGATCATCCCCGAACAGCCTGCACCACCGGCACCCCTACCATTGCCTCTGCCTCCATTGGAGGACCCCACT ACGGATGCACTTGCCGGAACTGGCGAAGAAGGTGATGACACCGACCGTGAAGGCGATCCAGAGCCCGAGACAGTC GCAACCCCTGGAAGGAAGAAGCCGAGGAGAGGTAAACGAGGAAAGAAGAAGAAGAAGGACGCCACGCCTATCCCT GGTGAAGAAGGTGTTGAGGGCAAGGCGCCTAATGGACCCGATGGACCAGCTGCGAATGGCACACCAAACATCAAT CCGCCAGACAACAACCCAGATGCCACTATCGCTCCTGCAGCCCCACAATCTCCTTCACTTGTCGTCTCATCTCCC ATAATTCCTGCGGCCGCAGGTCCATCGTTAGTCGTGTCCGATACCATTCTCGGCTTTGGCTCTCATGGAACTGTT GTCTTCCAAGGTTCTCTACAGGGTCGAGCAGTCGCAGTGAAGCGGCTCCTCAAAGACTTTGTTACTCTGGCTTCA CGTGAAGTCAGCATCCTCCAAGAGAGTGACGACCACCCGAATGTTATCCGATATTACTACCAAGAAGCCCACGCC AATTTCCTCTATATCGCTCTTGAACTCTGCCCCGCATCCCTAGCCGACATCATCGAAACTCCTGACCGAGAAGCC TTCAGAGATATCGCCATTTCCTTCGACCCCAAGAGGGCGTTGAAGCAGATCACGAGCGGTCTCAAGCACCTCCAT GCGCTCAAACTCGTCCACCGTGACATCAAGCCCCAGAATATTCTTATTACGACAAACAAGAGCACTGGACGTGGG CGACCGACATATCGTATGCTCATCTCTGACTTCGGATTGTGCAAGAAGTTGGACGTTGACCAAACGAGTTTCTTA CCAACCATGAATGGCGGGATGGGTGCTGGGACGGTCGGGTGGAGAGCACCGGAGATTTTACGTGGCGAGGTCAAG CTTGATGATTTGAGCGATGATCACTCCATGAGTTCGAGAGGAAGTGTGTCAACCATTAACGGGTCATCATCGTCG TCGTCGACGTCGGGTCTCTCAACCGCGAAGCCAACACGACTCACCAAGTCTGTGGATATCTTCGCCCTTGGTTGC CTGTACTACTACACCTTGACCAACGGCAGCCATCCTTACGGGGATCGATTCGAGCGGGAAGTCAACATCTTGAAG GACGCCAAGTCGCTGGATTTACTGGAGAGATTTGGAGAGGAAGGAACCGAAGCGTGCGATTTGATCGAGAAGATG TTGGATCCCGAGGCATCTGAAAGACCGGATACGACTGCGTGCCTATTGCACCCCTTCTTCTGGGATCCCGCACGC CGCCTTAACTTCCTCCAAGACGCCTCGGACCGGTTCGAAATCATGTGTCGGGACCCCAAGGATCCTCTCCTACTG CAACTCGAAACCGGTGCGCTGGCCATCGTTGGAAATGACTGGCATGCAAGACTGGACAGGTTGTTTGTTGAGAAC CTTGGCAAGTTTAGAAAGTATGACGGAAAGTCTGTACAAGATTTGCTTCGTGCTCTTCGTAACAAGAAACACCAT TATCAAGACTTGCCTGACAATGTTAAGCGAAGTCTTGGGCCGATGCCAGAGGGCTTCCTGGCCTACTTCACGAAA CGTTTCCCCCGCCTCTTCTTACATGTCCATCGTGTCATCTCCGACACTGTACTACGGACAGAGTCTATGTTTAGA TCTTACTTTGAACTGCCTGACTCCTGA |
| Length | 3477 |
Transcript
| Sequence id | CopciAB_364173.T0 |
|---|---|
| Sequence |
>CopciAB_364173.T0 GGTTTTCTGCCACCACCACCACCACGAACGACGACTAGAAACCAGCGCCCAACGCCGGGCTGAACGCTTATGGCG TTCGTATTCTTCCTACAGTTCTTGCTCCTCATCGTGGGCGCGGTCTGCTGGGCAGAGATCGTCAATGTCCGGTCA ACCATCCCAGCAGCGGAGACGGCGGCGCTCATACAACGAACGAGACCAGCACCTACTTATGCGACCGGAGGGATT CTAGAACCCACCAAAGACCTAGAGCTGCTGGACATTGTCCTGGTAGCCTCGATCGACGGCAAATTCCATGCACTC AGTCGCTCGACGGGCAAGATCTTGTGGTCTATGCCTGCATTCGCACGGGCAGAAGGCTCGTCAACTCTTTCAAGC TTGGCGCCTCTGGTTAGGACGACACACCCAGAAAACGACCCCGACGACTTTGACGACGATACAACACCAGAAACC TATATCATTGAACCGCAGTCTGGCCACATCTATGTCGTAAACTCAGAAGACGACTCGTTGCAACGGTTCCCGTTC TCAATGGCGGAGTTGGTTGACATGTCGCCGTTCACCTCCCCAGGTGACGACGAGAAGAGGGTATTCATTGGGCGC AAGGAGACTTCACTATTGTTACTGGAGCTCGAAACAGGAAGAATCAAGGCGACCCTCAACTCCGAATGTCCGCCT TATGACGACAGGCGTATTGACCTCGACGAGCTGGAAGAACCAGAGTCGCGGGGCTCATCGACCACCGAAGTATTC ATTGGGCGAACAGATTATTACATTTCCATCTATCCCCACCGCTCTAAGGGCTACGGCACTCGGGCGCCCGTCCAA AACCTGTCATTCTCAGTCTACGGACCCAACAACGAAGACAACTTGCTCCAAGCAAACTATCGAAGGACTAGCGAC GATGCGTACATCCAAGGTTTTCCCAACGGGGATGTCTACTCCTTTAAGACCAAGTCGGAAGGGGGTAGAGTCCAA ACACAACCTTTATGGGTTCAGAAATTCCATGCACCGACGGTCGCTGTTTTTGATGTCCTACGGGGCTCAACTCCA CTTCAGCCACATCCTCACGCCTTTGTTCTCCTGCAACCACGACCACGCGCGGAAGATGTCATCCCAGGTCTCGAC CGCTCTCTCTTACGTGGTGCAAAGTCGGCCTACGTTGGGATGGTGGAGGAAACGGGGAGCCTTTACGCTATGAGC CCTTCACATTATCCACTGGTTGCGTTTGGGGACGCAGAGGAGGGCAGAAAGGGACGACTTATTGAGGGTTCTGGG GGTTCAGCGCAAAGCATCCATGATATCATCCGCAACCAGAAGGAGGCTGACAGGCGGATTAGATGCGAGGATAAC CCGAGAGACCGCGAGTGTCTACTGGGTATGCATCCGCTTGACGAAACGGACGGCCATGAAAGTCGAATGCGACGA CTGCTCGAAGGACCTTCAAGCATCAGCCCCATCCCGACGTTCAGACCCGGTCAGAACCAATGGATCGATCACGGA ATCGAAGTCGTCCCCAACATTGAAGCTCCACCCAACAATTCTCAACAACCTGCCACGCCTGTCCCCCCGGAGGAA GTACGGAAGACGGGATGGGAATATCTGGGAGTAACACTCGGGCTGGGTTTGGTGTCTTTGTGGTTCTTATTCAAG AAGTTCGGCCAGCGAGAGGGATTCAAGGTACCCAAATTCATTCCCGTACCCAACGGGGACGCGCAGATCGCCCGT CCCCTTGCTCCCATACCGGACACTGCCACCGAGCCAGCACCAGCACCCGAACCAGCTACGGTCCAGCCTCTTCAG ATCATCCCCGAACAGCCTGCACCACCGGCACCCCTACCATTGCCTCTGCCTCCATTGGAGGACCCCACTACGGAT GCACTTGCCGGAACTGGCGAAGAAGGTGATGACACCGACCGTGAAGGCGATCCAGAGCCCGAGACAGTCGCAACC CCTGGAAGGAAGAAGCCGAGGAGAGGTAAACGAGGAAAGAAGAAGAAGAAGGACGCCACGCCTATCCCTGGTGAA GAAGGTGTTGAGGGCAAGGCGCCTAATGGACCCGATGGACCAGCTGCGAATGGCACACCAAACATCAATCCGCCA GACAACAACCCAGATGCCACTATCGCTCCTGCAGCCCCACAATCTCCTTCACTTGTCGTCTCATCTCCCATAATT CCTGCGGCCGCAGGTCCATCGTTAGTCGTGTCCGATACCATTCTCGGCTTTGGCTCTCATGGAACTGTTGTCTTC CAAGGTTCTCTACAGGGTCGAGCAGTCGCAGTGAAGCGGCTCCTCAAAGACTTTGTTACTCTGGCTTCACGTGAA GTCAGCATCCTCCAAGAGAGTGACGACCACCCGAATGTTATCCGATATTACTACCAAGAAGCCCACGCCAATTTC CTCTATATCGCTCTTGAACTCTGCCCCGCATCCCTAGCCGACATCATCGAAACTCCTGACCGAGAAGCCTTCAGA GATATCGCCATTTCCTTCGACCCCAAGAGGGCGTTGAAGCAGATCACGAGCGGTCTCAAGCACCTCCATGCGCTC AAACTCGTCCACCGTGACATCAAGCCCCAGAATATTCTTATTACGACAAACAAGAGCACTGGACGTGGGCGACCG ACATATCGTATGCTCATCTCTGACTTCGGATTGTGCAAGAAGTTGGACGTTGACCAAACGAGTTTCTTACCAACC ATGAATGGCGGGATGGGTGCTGGGACGGTCGGGTGGAGAGCACCGGAGATTTTACGTGGCGAGGTCAAGCTTGAT GATTTGAGCGATGATCACTCCATGAGTTCGAGAGGAAGTGTGTCAACCATTAACGGGTCATCATCGTCGTCGTCG ACGTCGGGTCTCTCAACCGCGAAGCCAACACGACTCACCAAGTCTGTGGATATCTTCGCCCTTGGTTGCCTGTAC TACTACACCTTGACCAACGGCAGCCATCCTTACGGGGATCGATTCGAGCGGGAAGTCAACATCTTGAAGGACGCC AAGTCGCTGGATTTACTGGAGAGATTTGGAGAGGAAGGAACCGAAGCGTGCGATTTGATCGAGAAGATGTTGGAT CCCGAGGCATCTGAAAGACCGGATACGACTGCGTGCCTATTGCACCCCTTCTTCTGGGATCCCGCACGCCGCCTT AACTTCCTCCAAGACGCCTCGGACCGGTTCGAAATCATGTGTCGGGACCCCAAGGATCCTCTCCTACTGCAACTC GAAACCGGTGCGCTGGCCATCGTTGGAAATGACTGGCATGCAAGACTGGACAGGTTGTTTGTTGAGAACCTTGGC AAGTTTAGAAAGTATGACGGAAAGTCTGTACAAGATTTGCTTCGTGCTCTTCGTAACAAGAAACACCATTATCAA GACTTGCCTGACAATGTTAAGCGAAGTCTTGGGCCGATGCCAGAGGGCTTCCTGGCCTACTTCACGAAACGTTTC CCCCGCCTCTTCTTACATGTCCATCGTGTCATCTCCGACACTGTACTACGGACAGAGTCTATGTTTAGATCTTAC TTTGAACTGCCTGACTCCTGATCGCATTTCGCTTAGTTCCTGTACATTCTCCCTCCCATCCTCACTTCCCGCGAT ACAGTAATTCATCGACCGACCCGACGGTTATTGCATTCTCTTCAAAAGTTGACGTACTTACCGCTTATATCCTAG CTCAACTCATCCGCATTGCTCAAGTACTACAATCCTGCTAGTTATCTCACTGCATATTTATCTTGATGCCTCTCA AACGTGTATTATCTGTACTTCTAAGTAGTTAGCTTTCTGATGCCCCTGCCTGTGTTACATATGATACCGCCAATG GAATTTCTTTGCACATAG |
| Length | 3843 |
Gene
| Sequence id | CopciAB_364173.T0 |
|---|---|
| Sequence |
>CopciAB_364173.T0 GGTTTTCTGCCACCACCACCACCACGAACGACGACTAGAAACCAGCGCCCAACGCCGGGCTGAACGCTTATGGCG TTCGTATTCTTCCTACAGTTCTTGCTCCTCATCGTGGGCGCGGTCTGCTGGGCAGAGATCGTCAATGTCCGGTCA ACCATCCCAGCAGCGGAGACGGCGGCGCTCATACAACGAACGAGACCAGCACCTACTTATGCGACCGGAGGGATT CTAGAACCCACCAAAGACCTAGAGCTGCTGGACATTGTCCTGGTAGCCTCGATCGACGGCAAATTCCATGCACTC AGTCGCTCGACGGGCAAGATCTTGTGGTCTATGCCTGCATTCGCACGGGCAGAAGGCTCGTCAACTCTTTCAAGC TTGGCGCCTCTGGTTAGGACGACACACCCAGAAAACGACCCCGACGACTTTGACGACGATACAACACCAGAAACC TATATCATTGAACCGCAGTCTGGCCACATCTATGTCGTAAACTCAGAAGACGACTCGTTGCAACGGTTCCCGTTC TCAATGGCGGAGTTGGTTGACATGTCGCCGTTCACCTCCCCAGGTGACGACGAGAAGAGGGTATTCATTGGGCGC AAGGAGACTTCACTATTGTTACTGGAGCTCGAAACAGGAAGAATCAAGGCGACCCTCAACTCCGAATGTCCGCCT TATGACGACAGGCGTATTGACCTCGACGAGCTGGAAGAACCAGAGTCGCGGGGCTCATCGACCACCGAAGTATTC ATTGGGCGAACAGGTGCGGTGTTGACAGATTTGCGACCTGGAATTCAGCGCTGATGGCTAATTTCCCTACTTGGC AGATTATTACATTTCCATCTATCCCCACCGCTCTAAGGGCTACGGCACTCGGGCGCCCGTCCAAAACCTGTCATT CTCAGTCTACGGACCCAACAACGAAGACAACTTGCTCCAAGCAAACTATCGAAGGACTAGCGACGATGCGTACAT CCAAGGTTTTCCCAACGGGGATGTCTACTCCTTTAAGACCAAGTCGGAAGGGGGTAGAGTCCAAACACAACCTTT ATGGGTTCAGAAATTCCATGCACCGACGTGAGTTTGACGTTAAGAGTACCCTCGTTACATACTCAACAGCTTGCA GGGTCGCTGTTTTTGATGTCCTACGGGGCTCAACTCCACTTCAGCCACATCCTCACGCCTTTGTTCTCCTGCAAC CACGACCACGCGCGGAAGATGTCATCCCAGGTCTCGACCGCTCTCTCTTACGTGGTGCAAAGTCGGCCTACGTTG GGATGGTGGAGGAAACGGGGAGCCTTTACGCTATGAGCCCTTCACATTATCCACTGGTTGCGTTTGGGGACGCAG AGGAGGGCAGAAAGGGACGACTTATTGAGGGTTCTGGGGGTTCAGCGCAAAGCATCCATGATATCATCCGCAACC AGAAGGAGGCTGACAGGCGGATTAGATGCGAGGATAACCCGAGAGACCGCGAGTGTCTACTGGGTATGCATCCGC TTGACGAAACGGACGGCCATGAAAGTCGAATGCGACGACTGCTCGAAGGACCTTCAAGCATCAGCCCCATCCCGA CGTTCAGACCCGGTCAGAACCAATGGATCGATCACGGAATCGAAGTCGTCCCCAACATTGAAGCTCCACCCAACA ATTCTCAACAACCTGCCACGCCTGTCCCCCCGGAGGAAGTACGGAAGACGGGATGGGAATATCTGGGAGTAACAC TCGGGCTGGGTTTGGTGTCTTTGTGGTTCTTATTCAAGAAGTTCGGCCAGCGAGAGGGATTCAAGGTACCCAAAT TCATTCCCGTACCCAACGGGGACGCGCAGATCGCCCGTCCCCTTGCTCCCATACCGGACACTGCCACCGAGCCAG CACCAGCACCCGAACCAGCTACGGTCCAGCCTCTTCAGATCATCCCCGAACAGCCTGCACCACCGGCACCCCTAC CATTGCCTCTGCCTCCATTGGAGGACCCCACTACGGATGCACTTGCCGGAACTGGCGAAGAAGGTGATGACACCG ACCGTGAAGGCGATCCAGAGCCCGAGACAGTCGCAACCCCTGGAAGGAAGAAGCCGAGGAGAGGTAAACGAGGAA AGAAGAAGAAGAAGGACGCCACGCCTATCCCTGGTGAAGAAGGTGTTGAGGGCAAGGCGCCTAATGGACCCGATG GACCAGCTGCGAATGGCACACCAAACATCAATCCGCCAGACAACAACCCAGATGCCACTATCGCTCCTGCAGCCC CACAATCTCCTTCACTTGTCGTCTCATCTCCCATAATTCCTGCGGCCGCAGGTCCATCGTTAGTCGTGTCCGATA CCATTCTCGGTGAGCACTGCCCTTCCTGTCACTGTCAATCAATTGAACTGACTTCTTCTTAGGCTTTGGCTCTCA TGGAACTGTTGTCTTCCAAGGTTCTCTACAGGGTCGAGCAGTCGCAGTGAAGCGGCTCCTCAAAGACTTTGTTAC TCTGGCTTCACGTGAAGTCAGCATCCTCCAAGAGAGTGACGACCACCCGAATGTTATCCGATATTACTACCAAGA AGCCCACGCCAATTTCCTCTATATCGCTCTTGAACTCTGCCCCGCATCCCTAGCCGACATCATCGAAACTCCTGA CCGAGAAGCCTTCAGAGATATCGCCATTTCCTTCGACCCCAAGAGGGCGTTGAAGCAGATCACGAGCGGTCTCAA GCACCTCCATGCGCTCAAACTCGTCCACCGTGACATCAAGCCCCAGAATATTCTTATTACGACAAACAAGAGCAC TGGACGTGGGCGACCGACATATCGTATGCTCATCTCTGACTTCGGATTGTGCAAGAAGTTGGACGTTGACCAAAC GAGTTTCTTACCAACCATGAATGGCGGGATGGGTGCTGGGACGGTCGGGTGGAGAGCACCGGAGATTTTACGTGG CGAGGTCAAGCTTGATGATTTGAGCGATGATCACTCCATGAGTTCGAGAGGAAGTGTGTCAACCATTAACGGGTC ATCATCGTCGTCGTCGACGTCGGGTCTCTCAACCGCGAAGCCAACACGACTCACCAAGTCTGTGGATATCTTCGC CCTTGGTTGCCTGTACTACTACACCTTGACCAACGGCAGCCATCCTTACGGGGATCGATTCGAGCGGGAAGTCAA CATCTTGAAGGACGCCAAGTCGCTGGATTTACTGGAGAGATTTGGAGAGGAAGGAACCGAAGCGTGCGATTTGAT CGAGAAGATGTTGGATCCCGAGGCATCTGAAAGGTATGCCAGCTTTGATACTCTAATTGCCTATTTTCGGACGGA ACGCCTAACCCTTGATCATCAGACCGGATACGACTGCGTGCCTATTGCACCCCTTCTTCTGGGATCCCGCACGCC GCCTTAACTTCCTCCAAGACGCCTCGGACCGGTTCGAAATCATGTGTCGGGACCCCAAGGATCCTCTCCTACTGC AACTCGAAACCGGTGCGCTGGCCATCGTTGGAAATGACTGGCATGCAAGACTGGACAGGTTGTTTGTTGAGAACC TTGGCAAGTTTAGAAAGTATGACGGAAAGTCTGTACAAGATTTGCTTCGTGCTCTTCGTAACAAGGTATGCTGTT TCCGCCTTGATGCCTCCTCCAATGGATACTCACATCTCATTTGTAGAAACACCATTATCAAGACTTGCCTGACAA TGTTAAGCGAAGTCTTGGGCCGATGCCAGAGGGCTTCCTGGCCTACTTCACGAAACGTTTCCCCCGCCTCTTCTT ACATGTCCATCGTGTCATCTCCGACACTGTACTACGGACAGAGTCTATGTTTAGATCTTACTTTGAACTGCCTGA CTCCTGATCGCATTTCGCTTAGTTCCTGTACATTCTCCCTCCCATCCTCACTTCCCGCGATACAGTAATTCATCG ACCGACCCGACGGTTATTGCATTCTCTTCAAAAGTTGACGTACTTACCGCTTATATCCTAGCTCAACTCATCCGC ATTGCTCAAGTACTACAATCCTGCTAGTTATCTCACTGCATATTTATCTTGATGCCTCTCAAACGTGTATTATCT GTACTTCTAAGTAGTTAGCTTTCTGATGCCCCTGCCTGTGTTACATATGATACCGCCAATGGAATTTCTTTGCAC ATAG |
| Length | 4129 |