CopciAB_365598
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_365598 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 365598 |
| Uniprot id | Functional description | Domain of unknown function | |
| Location | scaffold_5:1094583..1097582 | Strand | + |
| Gene length (nt) | 3000 | Transcript length (nt) | 2794 |
| CDS length (nt) | 2508 | Protein length (aa) | 835 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7260064 | 53.9 | 9.27E-270 | 844 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB19923 | 52.4 | 8.171E-263 | 824 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1055497 | 47.9 | 2.656E-221 | 703 |
| Pleurotus ostreatus PC9 | PleosPC9_1_47322 | 47.8 | 5.736E-221 | 702 |
| Schizophyllum commune H4-8 | Schco3_2623257 | 46 | 2.444E-215 | 686 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1442943 | 46.2 | 2.329E-214 | 683 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_188624 | 44.3 | 2.11E-211 | 674 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_119933 | 43.8 | 5.198E-210 | 670 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_18801 | 45.9 | 1.082E-202 | 649 |
| Lentinula edodes NBRC 111202 | Lenedo1_1097137 | 45.8 | 1.166E-201 | 646 |
| Lentinula edodes B17 | Lened_B_1_1_11470 | 45.3 | 2.839E-196 | 630 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_64662 | 43.7 | 1.945E-192 | 619 |
| Flammulina velutipes | Flave_chr11AA00876 | 42.7 | 3.929E-181 | 586 |
| Grifola frondosa | Grifr_OBZ76602 | 44.2 | 3.192E-157 | 516 |
| Auricularia subglabra | Aurde3_1_1237497 | 51 | 9.08E-75 | 271 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_365598.T0 |
| Description | Domain of unknown function |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF12330 | Haspin like kinase domain | - | 453 | 736 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011009 | Protein kinase-like domain superfamily |
| IPR024604 | Serine/threonine-protein kinase haspin, C-terminal |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| K16315 |
EggNOG
| COG category | Description |
|---|---|
| D | Domain of unknown function |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
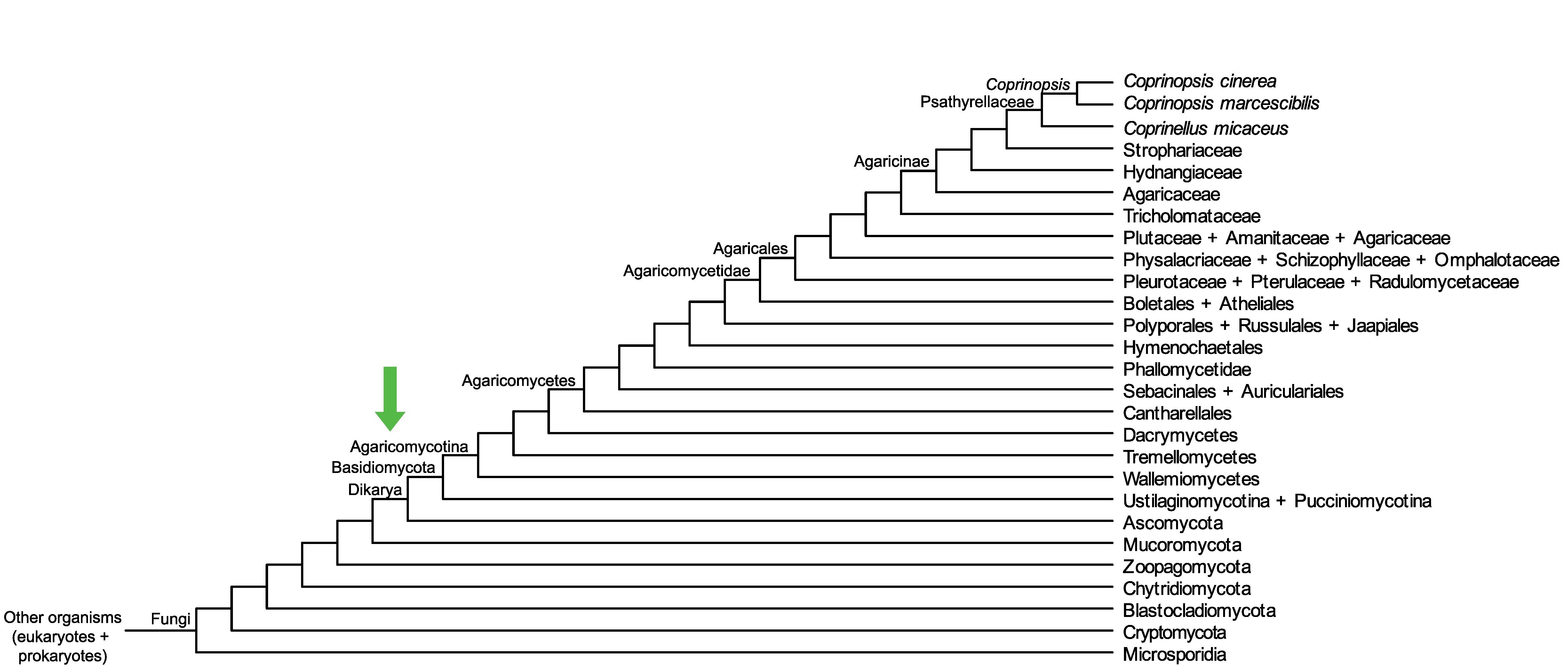
Conservation of CopciAB_365598 across fungi.
Arrow shows the origin of gene family containing CopciAB_365598.
Protein
| Sequence id | CopciAB_365598.T0 |
|---|---|
| Sequence |
>CopciAB_365598.T0 MLSAQTKQVNAYGKRARRVVNVTASSSGPAPGGIISIFDDLPPAPALMPLASRMKKRENIAPKPKSQLSPKPVGG KQRKRRLSPVLTPVKKVPRVAQLIKESRTQPLDNKLDGGVRQANAKVNLTTEAEASPFDASSFSSPRRPFSSFSV NVPGSPALPASLRASKSRVPSAKSIPFKLAKPSSPVVNMDIIVLDDDGQVVRKERRSSKTGVETNPVKDVPKKRA RPSRAAASKAAILVESDSDPDVARQPAQPKRTQRQKALVVVSDVSDSETENAPTAPSPAPPKTNRLSKLPTVEVV IPPAPYPIRPLARPSPPAGTQHRAATPTKPKPAPPTARPGPAFQLPPSPIPRARPLTPIKGMRRNGLFDPPSPPS PSLTDFDLSLDLSELNIDAECVDAHDVGPNIPAYLLPLLEECHQEECGPHDFSSFIDSFPFDPILRDGRSDAPSE LEFRKIGEASYSEVFGIADVVLKVIPLRDESAPGAISTELEEGPPPSDAQDVRKEIIVTRAMGEVYGGFVKFLKT YIVRGKYPELLLNLWDEYNETKGSESMRPDTFKLSQVYAIIVLPNGGPDLEAYSFNNPTKQGWKQACSIFWQVAK SLAHAEHLVSFEHRDLHWGQILVKNVPSQKPKLQPINVNGVKSRKQRLMMDDKSHGVQTTIIDLGLSRMDAGDGS GGYRVQWTPFDEEVFMGEGEYQFDVYRMMRDHTGDDWEGYHPLTNVMWLHYLATKLLYGKGLKAPVVRKPKSGSE APLATSSDAFSERDCYESLLDIEQWLGSFLTGAFPAQSKAIAKTKGRRKTIVLSSCLALRDGPSCAGEVVAYGVK KGWVSATKLF |
| Length | 835 |
Coding
| Sequence id | CopciAB_365598.T0 |
|---|---|
| Sequence |
>CopciAB_365598.T0 ATGCTCAGCGCACAGACAAAGCAGGTCAATGCATATGGCAAGCGGGCCAGGCGCGTTGTGAACGTTACCGCCAGC TCAAGTGGGCCAGCACCTGGCGGGATCATCAGCATATTCGACGATCTTCCGCCAGCACCCGCATTGATGCCTCTA GCATCGCGCATGAAGAAGCGGGAAAACATCGCTCCTAAACCAAAATCCCAACTTTCCCCCAAACCTGTCGGCGGA AAGCAGCGCAAGAGACGACTCTCGCCGGTCCTCACGCCTGTGAAGAAAGTACCACGAGTCGCCCAGCTGATCAAG GAGTCTCGAACTCAGCCCTTAGACAACAAATTGGACGGTGGCGTACGGCAAGCCAATGCTAAAGTTAATCTGACA ACGGAAGCAGAGGCTTCGCCGTTTGATGCAAGTTCATTTTCGTCTCCTCGCCGTCCTTTCTCAAGTTTCTCGGTC AATGTTCCCGGGTCTCCTGCTCTTCCTGCTAGCTTGCGCGCCTCCAAGTCACGCGTTCCCTCGGCCAAGAGCATT CCGTTTAAGCTGGCGAAGCCTAGCAGTCCGGTCGTAAACATGGACATTATCGTCCTCGACGATGATGGTCAAGTC GTTCGCAAAGAGAGAAGGTCCTCCAAGACAGGCGTTGAAACTAACCCTGTGAAGGATGTCCCGAAGAAGAGAGCT CGTCCATCCAGAGCAGCTGCCTCCAAAGCAGCAATCCTCGTAGAGTCCGACAGTGACCCTGATGTGGCTCGCCAA CCTGCCCAACCTAAACGCACACAACGGCAGAAAGCACTAGTTGTCGTCTCTGACGTGTCAGACTCAGAGACAGAA AATGCCCCAACTGCACCTTCCCCTGCCCCGCCGAAAACCAATCGACTCTCGAAGCTTCCTACGGTGGAAGTAGTG ATACCCCCCGCACCTTATCCTATACGACCACTCGCGAGGCCCTCTCCACCAGCGGGTACGCAACATCGGGCTGCA ACACCGACGAAACCCAAACCCGCCCCACCTACTGCACGGCCAGGGCCCGCTTTCCAGCTACCACCCTCCCCCATA CCGAGAGCTCGCCCGCTAACCCCCATCAAGGGCATGCGGCGGAATGGCCTATTTGATCCACCTTCTCCCCCATCC CCTTCACTCACAGACTTTGACTTGTCTCTGGATCTCTCCGAGCTCAATATCGACGCCGAATGTGTTGATGCTCAC GATGTTGGACCAAACATTCCCGCTTACCTGCTTCCACTCCTGGAAGAGTGTCATCAAGAGGAATGCGGACCTCAT GACTTTTCCTCCTTCATTGACAGCTTTCCGTTCGATCCTATTCTCCGCGATGGGCGGTCTGATGCGCCGAGCGAG TTGGAATTCAGGAAGATTGGGGAGGCTAGCTACTCGGAAGTCTTTGGTATAGCCGATGTCGTCCTCAAGGTCATT CCGTTGCGCGACGAGTCTGCCCCAGGAGCAATCTCAACGGAGCTGGAGGAAGGACCTCCTCCAAGTGATGCGCAG GACGTCCGCAAGGAAATCATCGTCACCAGGGCCATGGGCGAAGTCTACGGTGGATTTGTGAAATTCTTGAAGACT TATATCGTCAGGGGAAAGTATCCGGAACTTCTTCTCAACCTTTGGGATGAATATAACGAAACGAAGGGATCAGAG AGTATGCGACCAGACACGTTCAAACTCTCTCAAGTCTACGCTATCATCGTACTGCCCAACGGTGGCCCTGACCTC GAGGCTTACTCGTTTAACAACCCTACCAAACAGGGATGGAAGCAAGCCTGCAGCATCTTCTGGCAAGTGGCCAAA TCACTCGCTCACGCGGAGCACCTGGTCTCGTTTGAGCATCGCGACCTGCACTGGGGTCAGATACTGGTGAAAAAT GTCCCGTCACAGAAGCCCAAACTCCAACCTATCAACGTGAATGGAGTCAAATCTAGGAAGCAACGCCTCATGATG GACGACAAGTCCCATGGGGTCCAAACAACCATCATTGACTTGGGCCTCTCGCGCATGGACGCTGGAGATGGATCA GGTGGATATCGTGTGCAATGGACACCATTCGACGAGGAGGTTTTCATGGGCGAAGGCGAATACCAGTTCGATGTT TATCGGATGATGAGGGACCACACTGGAGATGATTGGGAAGGTTACCACCCTTTGACCAATGTGATGTGGCTACAC TACCTTGCAACCAAACTTCTCTATGGCAAAGGGCTCAAGGCGCCGGTGGTGCGCAAACCGAAATCCGGGTCTGAA GCGCCTCTGGCAACCTCCTCCGACGCTTTCTCAGAAAGGGACTGCTACGAGTCGTTGCTTGATATTGAGCAATGG TTGGGAAGTTTCTTGACCGGGGCATTCCCTGCACAGTCGAAGGCGATTGCCAAAACCAAGGGAAGGCGTAAGACA ATTGTCCTTTCCAGCTGTCTTGCCTTGCGCGATGGGCCTTCCTGTGCTGGCGAAGTCGTTGCCTACGGTGTCAAA AAGGGCTGGGTCAGTGCTACGAAATTATTCTAA |
| Length | 2508 |
Transcript
| Sequence id | CopciAB_365598.T0 |
|---|---|
| Sequence |
>CopciAB_365598.T0 GGTGCCTCTTTGCTCATCGCCACGACGCCAACAATATCGCCGTCTCAGCTTTTCTAGTACACGAAGCATCGCAGT TGCTGCTCTCGTTCTCAAGATGCTCAGCGCACAGACAAAGCAGGTCAATGCATATGGCAAGCGGGCCAGGCGCGT TGTGAACGTTACCGCCAGCTCAAGTGGGCCAGCACCTGGCGGGATCATCAGCATATTCGACGATCTTCCGCCAGC ACCCGCATTGATGCCTCTAGCATCGCGCATGAAGAAGCGGGAAAACATCGCTCCTAAACCAAAATCCCAACTTTC CCCCAAACCTGTCGGCGGAAAGCAGCGCAAGAGACGACTCTCGCCGGTCCTCACGCCTGTGAAGAAAGTACCACG AGTCGCCCAGCTGATCAAGGAGTCTCGAACTCAGCCCTTAGACAACAAATTGGACGGTGGCGTACGGCAAGCCAA TGCTAAAGTTAATCTGACAACGGAAGCAGAGGCTTCGCCGTTTGATGCAAGTTCATTTTCGTCTCCTCGCCGTCC TTTCTCAAGTTTCTCGGTCAATGTTCCCGGGTCTCCTGCTCTTCCTGCTAGCTTGCGCGCCTCCAAGTCACGCGT TCCCTCGGCCAAGAGCATTCCGTTTAAGCTGGCGAAGCCTAGCAGTCCGGTCGTAAACATGGACATTATCGTCCT CGACGATGATGGTCAAGTCGTTCGCAAAGAGAGAAGGTCCTCCAAGACAGGCGTTGAAACTAACCCTGTGAAGGA TGTCCCGAAGAAGAGAGCTCGTCCATCCAGAGCAGCTGCCTCCAAAGCAGCAATCCTCGTAGAGTCCGACAGTGA CCCTGATGTGGCTCGCCAACCTGCCCAACCTAAACGCACACAACGGCAGAAAGCACTAGTTGTCGTCTCTGACGT GTCAGACTCAGAGACAGAAAATGCCCCAACTGCACCTTCCCCTGCCCCGCCGAAAACCAATCGACTCTCGAAGCT TCCTACGGTGGAAGTAGTGATACCCCCCGCACCTTATCCTATACGACCACTCGCGAGGCCCTCTCCACCAGCGGG TACGCAACATCGGGCTGCAACACCGACGAAACCCAAACCCGCCCCACCTACTGCACGGCCAGGGCCCGCTTTCCA GCTACCACCCTCCCCCATACCGAGAGCTCGCCCGCTAACCCCCATCAAGGGCATGCGGCGGAATGGCCTATTTGA TCCACCTTCTCCCCCATCCCCTTCACTCACAGACTTTGACTTGTCTCTGGATCTCTCCGAGCTCAATATCGACGC CGAATGTGTTGATGCTCACGATGTTGGACCAAACATTCCCGCTTACCTGCTTCCACTCCTGGAAGAGTGTCATCA AGAGGAATGCGGACCTCATGACTTTTCCTCCTTCATTGACAGCTTTCCGTTCGATCCTATTCTCCGCGATGGGCG GTCTGATGCGCCGAGCGAGTTGGAATTCAGGAAGATTGGGGAGGCTAGCTACTCGGAAGTCTTTGGTATAGCCGA TGTCGTCCTCAAGGTCATTCCGTTGCGCGACGAGTCTGCCCCAGGAGCAATCTCAACGGAGCTGGAGGAAGGACC TCCTCCAAGTGATGCGCAGGACGTCCGCAAGGAAATCATCGTCACCAGGGCCATGGGCGAAGTCTACGGTGGATT TGTGAAATTCTTGAAGACTTATATCGTCAGGGGAAAGTATCCGGAACTTCTTCTCAACCTTTGGGATGAATATAA CGAAACGAAGGGATCAGAGAGTATGCGACCAGACACGTTCAAACTCTCTCAAGTCTACGCTATCATCGTACTGCC CAACGGTGGCCCTGACCTCGAGGCTTACTCGTTTAACAACCCTACCAAACAGGGATGGAAGCAAGCCTGCAGCAT CTTCTGGCAAGTGGCCAAATCACTCGCTCACGCGGAGCACCTGGTCTCGTTTGAGCATCGCGACCTGCACTGGGG TCAGATACTGGTGAAAAATGTCCCGTCACAGAAGCCCAAACTCCAACCTATCAACGTGAATGGAGTCAAATCTAG GAAGCAACGCCTCATGATGGACGACAAGTCCCATGGGGTCCAAACAACCATCATTGACTTGGGCCTCTCGCGCAT GGACGCTGGAGATGGATCAGGTGGATATCGTGTGCAATGGACACCATTCGACGAGGAGGTTTTCATGGGCGAAGG CGAATACCAGTTCGATGTTTATCGGATGATGAGGGACCACACTGGAGATGATTGGGAAGGTTACCACCCTTTGAC CAATGTGATGTGGCTACACTACCTTGCAACCAAACTTCTCTATGGCAAAGGGCTCAAGGCGCCGGTGGTGCGCAA ACCGAAATCCGGGTCTGAAGCGCCTCTGGCAACCTCCTCCGACGCTTTCTCAGAAAGGGACTGCTACGAGTCGTT GCTTGATATTGAGCAATGGTTGGGAAGTTTCTTGACCGGGGCATTCCCTGCACAGTCGAAGGCGATTGCCAAAAC CAAGGGAAGGCGTAAGACAATTGTCCTTTCCAGCTGTCTTGCCTTGCGCGATGGGCCTTCCTGTGCTGGCGAAGT CGTTGCCTACGGTGTCAAAAAGGGCTGGGTCAGTGCTACGAAATTATTCTAAGTGTCTCTCTGCTTTGCGATTTG GATAGACTCCTCTTATCACGGGACACCTGCATTTCTTGTATGCTCGTTTGCTTTCGTTTTCGTTCAAGGTATGGA CTTGTTTGTATGTACAACAAGTTTTGTAGAATTACATGGGGTACAATTGTGGTTCAACATCTTCAGTGCTGTACG TTGGGTCACCAACACTAGG |
| Length | 2794 |
Gene
| Sequence id | CopciAB_365598.T0 |
|---|---|
| Sequence |
>CopciAB_365598.T0 GGTGCCTCTTTGCTCATCGCCACGACGCCAACAATATCGCCGTCTCAGCTTTTCTAGTACACGAAGCATCGCAGT TGCTGCTCTCGTTCTCAAGATGCTCAGCGCACAGACAAAGCAGGTCAATGCATATGGCAAGCGGGCCAGGCGCGT TGTGAACGTTACCGCCAGCTCAAGTGGGCCAGCACCTGGCGGGATCATCAGCATATTCGACGATCTTCCGCCAGC ACCCGCATTGATGCCTCTAGCATCGCGCATGAAGAAGCGGGAAAACATCGCTCCTAAACCAAAATCCCAACTTTC CCCCAAACCTGTCGGCGGAAAGCAGCGCAAGAGACGACTCTCGCCGGTCCTCACGCCTGTGAAGAAAGTACCACG AGTCGCCCAGCTGATCAAGGAGTCTCGAACTCAGCCCTTAGACAACAAATTGGACGGTGGCGTACGGCAAGCCAA TGCTAAAGTTAATCTGACAACGGAAGCAGAGGCTTCGCCGTTTGATGCAAGTTCATTTTCGTCTCCTCGCCGTCC TTTCTCAAGTTTCTCGGTCAATGTTCCCGGGTCTCCTGCTCTTCCTGCTAGCTTGCGCGCCTCCAAGTCACGCGT TCCCTCGGCCAAGAGCATTCCGTTTAAGCTGGCGAAGCCTAGCAGTCCGGTCGTAAACATGGACATTATCGTCCT CGACGATGATGGTCAAGTCGTTCGCAAAGAGAGAAGGTCCTCCAAGACAGGCGTTGAAACTAACCCTGTGAAGGA TGTCCCGAAGAAGAGAGCTCGTCCATCCAGAGCAGCTGCCTCCAAAGCAGCAATCCTCGTAGAGTCCGACAGTGA CCCTGATGTGGCTCGCCAACCTGCCCAACCTAAACGCACACAACGGCAGAAAGCACTAGTTGTCGTCTCTGACGT GTCAGACTCAGAGACAGAAAATGCCCCAACTGCACCTTCCCCTGCCCCGCCGAAAACCAATCGACTCTCGAAGCT TCCTACGGTGGAAGTAGTGATACCCCCCGCACCTTATCCTATACGACCACTCGCGAGGCCCTCTCCACCAGCGGG TACGCAACATCGGGCTGCAACACCGACGAAACCCAAACCCGCCCCACCTACTGCACGGCCAGGGCCCGCTTTCCA GCTACCACCCTCCCCCATACCGAGAGCTCGCCCGCTAACCCCCATCAAGGGCATGCGGCGGAATGGCCTATTTGA TCCACCTTCTCCCCCATCCCCTTCACTCACAGACTTTGACTTGTCTCTGGATCTCTCCGAGCTCAATATCGACGC CGAATGTGTTGATGCTCACGATGTTGGACCAAACATTCCCGCTTACCTGCTTCCACTCCTGGAAGAGTGTCATCA AGAGGAATGCGGACCTCATGACTTTTCCTCCTTCATTGACAGCTTTCCGTTCGATCCTATTCTCCGCGATGGGCG GTCTGATGCGCCGAGCGAGTTGGAATTCAGGAAGATTGGGGAGGCTAGCTACTCGGAAGTCTTTGGTATAGCCGA TGTCGTCCTCAAGGTCATTCCGTTGCGCGACGAGTCTGCCCCAGGAGCAATCTCAACGGAGCTGGAGGAAGGACC TCCTCCAAGTGATGCGCAGGACGTCCGCAAGGAAATCATCGTCACCAGGGCCATGGGCGAAGTCTACGGTGGATT TGTGAAATTCTTGAAGACTTATATCGTCAGGGGAAAGTATCCGGAACTTCTTCTCAACCTTTGGGATGAATATAA CGAAACGAAGGGATCAGAGAGTATGCGACCAGGTACGGGTAGTCTTTTCGGAAAGGTGGAATTCACTAAGGTCCC CTTGCCAGACACGTTCAAACTCTCTCAAGTCTACGCTATCATCGTACTGCCCAACGGTGGCCCTGACCTCGAGGC TTACTCGTTTAACAACCCTACCAAACAGGGATGGAAGCAAGCCTGCAGCATCTTCTGGCAAGTGGCCAAATCACT CGCTCACGCGGAGCACCTGGTCTCGTTTGAGGTACGACCTGGCACGGCTTCCTGCATTCTCGTTCTGACTAGTCT TTCTAATAGCATCGCGACCTGCACTGGGGTCAGATACTGGTGAAAAATGTCCCGTCACAGAAGCCCAAACTCCAA CCTATCAACGTGAATGGAGTCAAATCTAGGAAGCAACGCCTCATGATGGACGACAAGTCCCATGGGGTCCAAACA ACCATCATTGACTTGGGCCTCTCGCGCATGGACGCTGGAGATGGATCAGGTGGATATCGTGTGCAATGGACACCA TTCGACGAGGAGGTTTTCATGGGCGAAGGTAAGCTTCCTCCGACTCTCGCCAAGGCAGTCACTGAAAGATGAATG GTAGGCGAATACCAGTTCGATGTTTATCGGATGATGAGGGACCACACTGGAGATGATTGGGAAGGTTACCACCCT TTGACCAATGTGATGGTAGGTACTTACTTTCTGGTCGTCGTGATTTATGACCAACTTTCTTGTTAGTGGCTACAC TACCTTGCAACCAAACTTCTCTATGGCAAAGGGCTCAAGGCGCCGGTGGTGCGCAAACCGAAATCCGGGTCTGAA GCGCCTCTGGCAACCTCCTCCGACGCTTTCTCAGAAAGGGACTGCTACGAGTCGTTGCTTGATATTGAGCAATGG TTGGGAAGTTTCTTGACCGGGGCATTCCCTGCACAGTCGAAGGCGATTGCCAAAACCAAGGGAAGGCGTAAGACA ATTGTCCTTTCCAGCTGTCTTGCCTTGCGCGATGGGCCTTCCTGTGCTGGCGAAGTCGTTGCCTACGGTGTCAAA AAGGGCTGGGTCAGTGCTACGAAATTATTCTAAGTGTCTCTCTGCTTTGCGATTTGGATAGACTCCTCTTATCAC GGGACACCTGCATTTCTTGTATGCTCGTTTGCTTTCGTTTTCGTTCAAGGTATGGACTTGTTTGTATGTACAACA AGTTTTGTAGAATTACATGGGGTACAATTGTGGTTCAACATCTTCAGTGCTGTACGTTGGGTCACCAACACTAGG |
| Length | 3000 |