CopciAB_369851
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_369851 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 369851 |
| Uniprot id | Functional description | Gmc oxidoreductase | |
| Location | scaffold_10:884119..886970 | Strand | - |
| Gene length (nt) | 2852 | Transcript length (nt) | 1941 |
| CDS length (nt) | 1827 | Protein length (aa) | 608 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Neurospora crassa | NCU09798 |
| Aspergillus nidulans | AN8329 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262709 | 55.4 | 2.897E-228 | 712 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB23961 | 55.8 | 7.227E-228 | 711 |
| Schizophyllum commune H4-8 | Schco3_2610771 | 43.8 | 2.754E-160 | 515 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_149638 | 45.7 | 4.919E-160 | 514 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1108891 | 44 | 3.726E-152 | 491 |
| Pleurotus ostreatus PC9 | PleosPC9_1_88910 | 44 | 3.647E-152 | 491 |
| Grifola frondosa | Grifr_OBZ73019 | 42.8 | 9.45E-151 | 487 |
| Lentinula edodes NBRC 111202 | Lenedo1_1236353 | 44.6 | 5.782E-148 | 479 |
| Flammulina velutipes | Flave_chr08AA00306 | 42.1 | 2.597E-147 | 477 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4827 | 43.7 | 1.307E-135 | 443 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1513595 | 39.2 | 4.606E-128 | 421 |
| Auricularia subglabra | Aurde3_1_1353711 | 37.8 | 2.659E-119 | 396 |
| Lentinula edodes B17 | Lened_B_1_1_14898 | 37.2 | 1.156E-107 | 361 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_120654 | 37.4 | 1.138E-106 | 358 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_131891 | 36.8 | 6.935E-102 | 344 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_369851.T0 |
| Description | Gmc oxidoreductase |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 15 | 326 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 462 | 599 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Gmc oxidoreductase |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
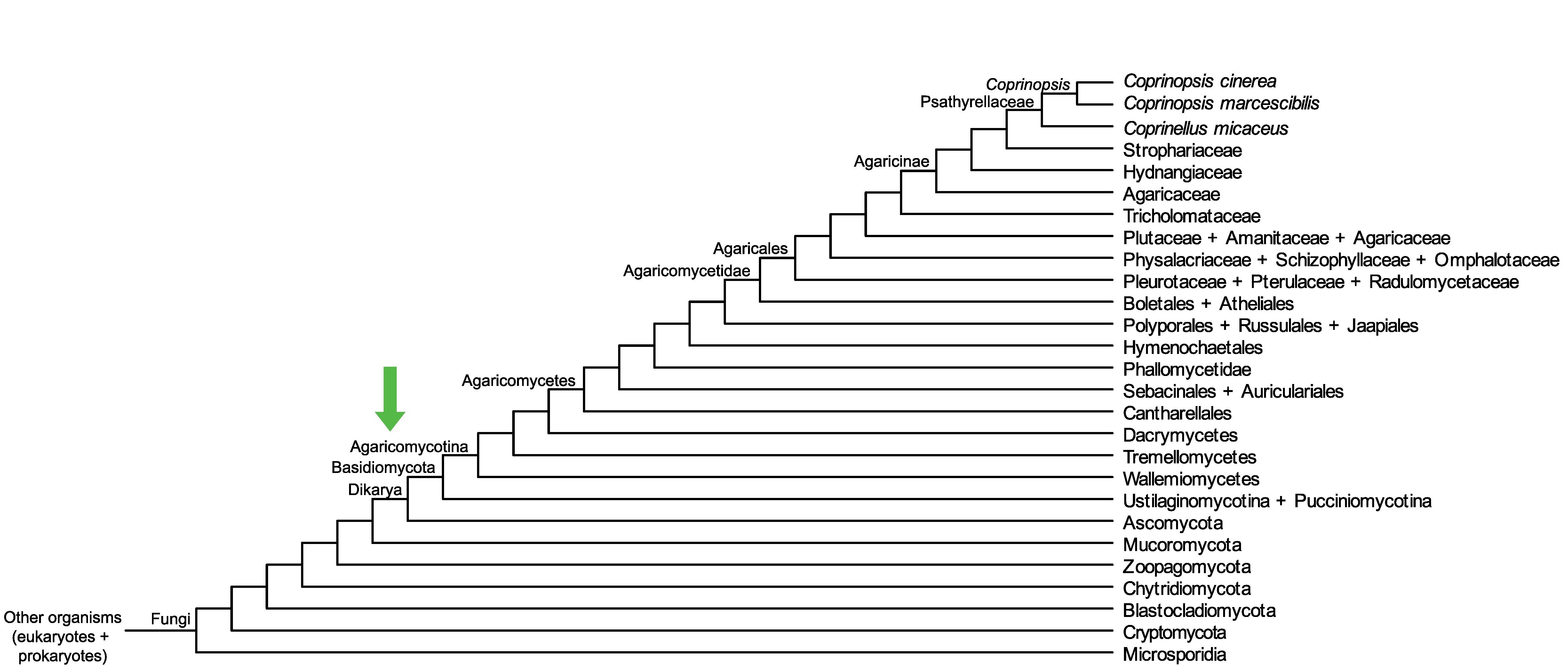
Conservation of CopciAB_369851 across fungi.
Arrow shows the origin of gene family containing CopciAB_369851.
Protein
| Sequence id | CopciAB_369851.T0 |
|---|---|
| Sequence |
>CopciAB_369851.T0 MPLIANPEEIANKTFDYVVVGAGTAGLTLATRLTEDENVSVLVLEAGIHHHGDPNIDVPAQYGATFMQPKYDWAF KTVKQKHSNDNEHVWARGKGLGGSSNMNFFCWIKPPAEDVDAIEKLGNPGWNWADYEAYSKKSETFHPAVPEIAE KYPHTYNAKFRGTSGPIQVTVPLHAHTLDEIWQNTLANKGVRKIEDPYGGDITGTWIAANNVDPKTWKRSYSATA YWEPNQHRPNLHVITEAYVTRIVWADEKDSEGNLVATGVQFVHGSTERTVLASKEVVLSAGTVKTPQILELSGVG KPEILSKFGIDTKLNLPGVGENVQEHHFMWVIYELDTTKGDHQTLDMFFEPGYVQESLKLHAEGKGLHRTGITSF AFLPLSSRDPKAAAALIDELEADLKKKKESGEISPALAKQYDIQLEILRNDKVPDMEFIVFPGYFMCPASQGTGR RFMTVISSLNHPISRGYIHIKSNNPLDDPEIDANYFGHNFDLKVLLEHVKYIRSLEDTEPFKSHTIKEVVPGPEC ASDADIENHIKNSHITTFHTVGSASMLPREDGGVVDPKLKVYGTKNLRVADLSIVPLHIAAHMQATAYVIGEKAA DLIKADAK |
| Length | 608 |
Coding
| Sequence id | CopciAB_369851.T0 |
|---|---|
| Sequence |
>CopciAB_369851.T0 ATGCCTCTCATCGCCAACCCTGAAGAAATTGCCAACAAGACCTTCGACTACGTCGTCGTCGGCGCAGGTACCGCT GGTTTGACCCTTGCCACTCGCCTCACCGAAGATGAGAATGTTAGTGTCCTCGTCCTCGAGGCCGGTATCCACCAC CACGGAGACCCCAACATCGATGTCCCGGCCCAATACGGTGCTACTTTCATGCAACCCAAGTACGACTGGGCCTTC AAGACTGTCAAGCAAAAACACTCCAACGACAATGAACACGTCTGGGCTCGTGGCAAAGGCCTTGGTGGAAGCTCA AACATGAACTTCTTCTGCTGGATCAAGCCTCCTGCGGAGGATGTCGATGCTATTGAGAAGCTTGGCAACCCTGGC TGGAACTGGGCCGATTATGAAGCTTACTCCAAGAAATCTGAGACGTTCCACCCTGCAGTCCCCGAGATTGCTGAG AAATACCCTCATACCTACAACGCGAAGTTCCGTGGAACCTCGGGGCCCATTCAAGTGACCGTACCCCTCCATGCC CACACGCTTGACGAGATCTGGCAAAACACACTCGCTAATAAGGGCGTGAGGAAAATCGAGGACCCCTATGGTGGG GATATCACGGGAACCTGGATCGCTGCTAACAACGTCGATCCGAAAACATGGAAACGCTCCTACTCCGCCACTGCC TACTGGGAGCCTAATCAACACCGTCCTAACCTTCACGTGATCACCGAGGCTTATGTTACCCGTATCGTCTGGGCG GATGAGAAGGATAGTGAGGGCAACCTGGTTGCTACTGGGGTCCAGTTCGTCCATGGCAGCACCGAAAGGACCGTT CTGGCGTCCAAGGAAGTCGTCTTGAGCGCTGGTACGGTGAAGACTCCCCAGATCCTCGAATTGTCCGGCGTCGGT AAACCAGAGATCCTCAGCAAGTTTGGTATCGATACCAAGTTGAACCTTCCGGGCGTCGGCGAGAACGTTCAGGAG CACCACTTCATGTGGGTGATCTACGAGTTGGACACTACCAAGGGTGACCATCAAACTCTCGATATGTTCTTCGAA CCTGGCTACGTCCAAGAATCCCTGAAGCTGCATGCCGAAGGCAAAGGTCTCCATCGTACCGGTATCACCTCCTTC GCCTTCTTGCCCTTATCGAGCAGAGACCCGAAGGCCGCGGCTGCCTTGATTGATGAGCTCGAAGCTGACCTCAAG AAGAAGAAGGAGAGCGGAGAGATCTCGCCTGCTCTTGCCAAGCAATATGACATCCAACTGGAGATTCTTCGCAAT GACAAGGTTCCTGACATGGAATTCATTGTGTTCCCTGGTTATTTCATGTGTCCCGCGAGCCAGGGTACTGGTCGA CGATTCATGACCGTCATCTCTTCTTTGAACCATCCAATCTCGAGAGGGTACATTCACATCAAGAGCAACAACCCC CTCGATGATCCTGAAATTGACGCCAACTACTTTGGTCACAACTTTGATCTCAAAGTTCTTTTGGAACACGTCAAG TACATTCGCTCTCTGGAAGATACCGAACCTTTCAAGTCGCACACCATCAAGGAGGTTGTTCCCGGTCCCGAATGC GCCAGCGACGCTGATATCGAGAACCACATCAAGAATTCGCATATTACCACTTTCCACACCGTTGGAAGCGCTTCG ATGTTGCCTCGAGAGGATGGTGGTGTCGTTGACCCCAAGCTCAAGGTCTATGGTACCAAGAACCTCCGCGTTGCT GACCTTTCCATCGTTCCCCTTCACATCGCTGCTCACATGCAAGCCACTGCCTATGTGATCGGAGAGAAAGCTGCT GATTTGATCAAGGCGGATGCGAAGTAA |
| Length | 1827 |
Transcript
| Sequence id | CopciAB_369851.T0 |
|---|---|
| Sequence |
>CopciAB_369851.T0 AGATATTCCACCAACTGCAACAACATGCCTCTCATCGCCAACCCTGAAGAAATTGCCAACAAGACCTTCGACTAC GTCGTCGTCGGCGCAGGTACCGCTGGTTTGACCCTTGCCACTCGCCTCACCGAAGATGAGAATGTTAGTGTCCTC GTCCTCGAGGCCGGTATCCACCACCACGGAGACCCCAACATCGATGTCCCGGCCCAATACGGTGCTACTTTCATG CAACCCAAGTACGACTGGGCCTTCAAGACTGTCAAGCAAAAACACTCCAACGACAATGAACACGTCTGGGCTCGT GGCAAAGGCCTTGGTGGAAGCTCAAACATGAACTTCTTCTGCTGGATCAAGCCTCCTGCGGAGGATGTCGATGCT ATTGAGAAGCTTGGCAACCCTGGCTGGAACTGGGCCGATTATGAAGCTTACTCCAAGAAATCTGAGACGTTCCAC CCTGCAGTCCCCGAGATTGCTGAGAAATACCCTCATACCTACAACGCGAAGTTCCGTGGAACCTCGGGGCCCATT CAAGTGACCGTACCCCTCCATGCCCACACGCTTGACGAGATCTGGCAAAACACACTCGCTAATAAGGGCGTGAGG AAAATCGAGGACCCCTATGGTGGGGATATCACGGGAACCTGGATCGCTGCTAACAACGTCGATCCGAAAACATGG AAACGCTCCTACTCCGCCACTGCCTACTGGGAGCCTAATCAACACCGTCCTAACCTTCACGTGATCACCGAGGCT TATGTTACCCGTATCGTCTGGGCGGATGAGAAGGATAGTGAGGGCAACCTGGTTGCTACTGGGGTCCAGTTCGTC CATGGCAGCACCGAAAGGACCGTTCTGGCGTCCAAGGAAGTCGTCTTGAGCGCTGGTACGGTGAAGACTCCCCAG ATCCTCGAATTGTCCGGCGTCGGTAAACCAGAGATCCTCAGCAAGTTTGGTATCGATACCAAGTTGAACCTTCCG GGCGTCGGCGAGAACGTTCAGGAGCACCACTTCATGTGGGTGATCTACGAGTTGGACACTACCAAGGGTGACCAT CAAACTCTCGATATGTTCTTCGAACCTGGCTACGTCCAAGAATCCCTGAAGCTGCATGCCGAAGGCAAAGGTCTC CATCGTACCGGTATCACCTCCTTCGCCTTCTTGCCCTTATCGAGCAGAGACCCGAAGGCCGCGGCTGCCTTGATT GATGAGCTCGAAGCTGACCTCAAGAAGAAGAAGGAGAGCGGAGAGATCTCGCCTGCTCTTGCCAAGCAATATGAC ATCCAACTGGAGATTCTTCGCAATGACAAGGTTCCTGACATGGAATTCATTGTGTTCCCTGGTTATTTCATGTGT CCCGCGAGCCAGGGTACTGGTCGACGATTCATGACCGTCATCTCTTCTTTGAACCATCCAATCTCGAGAGGGTAC ATTCACATCAAGAGCAACAACCCCCTCGATGATCCTGAAATTGACGCCAACTACTTTGGTCACAACTTTGATCTC AAAGTTCTTTTGGAACACGTCAAGTACATTCGCTCTCTGGAAGATACCGAACCTTTCAAGTCGCACACCATCAAG GAGGTTGTTCCCGGTCCCGAATGCGCCAGCGACGCTGATATCGAGAACCACATCAAGAATTCGCATATTACCACT TTCCACACCGTTGGAAGCGCTTCGATGTTGCCTCGAGAGGATGGTGGTGTCGTTGACCCCAAGCTCAAGGTCTAT GGTACCAAGAACCTCCGCGTTGCTGACCTTTCCATCGTTCCCCTTCACATCGCTGCTCACATGCAAGCCACTGCC TATGTGATCGGAGAGAAAGCTGCTGATTTGATCAAGGCGGATGCGAAGTAAAAGGCGAATGGATATATCGAAAAG AACAGGCAGTGTGTGTACATTATCTAGGGGTCGTTCTTTTCGGAACTGCAAGATATTTTTCGATTT |
| Length | 1941 |
Gene
| Sequence id | CopciAB_369851.T0 |
|---|---|
| Sequence |
>CopciAB_369851.T0 AGATATTCCACCAACTGCAACAACATGCCTCTCATCGCCAACCCTGAAGAAATTGCCAACAAGACCTTCGACTAC GTCGTCGTCGGCGCAGGTGTACGTTGTACTTTACACTCTCTGACCTTGGTCTTCCGTAACTCACCTAAAGTGATC GATTAGACCGCTGGTTTGACCCTTGCCACTCGCCTCACCGAAGATGAGAATGTTAGTGTCCTCGTCCTCGAGGCC GGTATCCACCACCACGGAGACCCCAACATCGATGTCCCGGCCCAATACGGTGCTACTTTCATGCAACCCAAGGTT AGCGCCCTCGATCCCATCACTCGATGTCCACCCCTTGACGTGTACCATAGTACGACTGGGCCTTCAAGACTGTCA AGCAAAAACACTCCAACGACAATGAACACGTCTGGGCTCGTGGCAAAGGCCTTGGTAAGTCGTCGTTGATCATCG CCTACTACGATTCTTACTCCTGTGGCAGGTGGAAGCTCAAACATGAACTTCTTCTGCTGGATCAAGCCTCCTGCG GAGGATGTCGATGCTATTGAGAAGCTTGGCAACCCTGGCTGGAACTGGGCCGATTATGAAGCTTACTCCAAGAAA TCTGAGACGTGGGTTATTCTGGTTCTGAACAACTGAATAGGATCTGGATCTAATGTTTGCATGCCAGGTTCCACC CTGCAGTCCCCGAGATTGCTGAGAAATACCCTCATACCTACAACGCGAAGTTCCGTGGAACCTCGGGGCCCATTC AAGTGACCGTACCCCTCCATGCCCACACGCTTGACGAGATCTGGCAAAACACACTCGCTAATAAGGGCGTGAGGA AAATCGAGGACCCCTATGGTGGGGATGTATGTGCTCCCATCGCGCTATTCTTAGTGTCCTTGAACTGACAAACTT TCACTCGGTGTAGATCACGGGAACCTGGATCGCTGCTAACAACGTCGATCCGAAAACATGGAAACGCTCCTACTC CGCCACTGCCTACTGGGAGCCTAATCAACACCGTCCTAACCTTCACGTATGTGCTCCAACCTTACGTTCGAGACA ATCCGCTTAGCCTGCTTGGAATTAAGGTGATCACCGAGGCTTATGTTACCCGTATCGTCTGGGCGGATGAGAAGG ATAGTGAGGGCAACCTGGTTGCTACTGGGGTCCAGTTCGTCCATGGCAGCACCGAAAGGACCGTTCTGGCGTCCA AGGAAGTCGTCTTGAGCGCTGGGTCAGTATTGGATCCGTGATGGTAATGGTGTTCTTTGATCTGATATGCTTTAT AGTACGGTGAAGACTCCCCAGATCCTCGAATTGTCCGGCGTCGGTAAACCAGAGATCCTCAGCAAGTTTGGTATC GATACCAAGTTGAACCTTCCGGGCGTCGGCGAGAACGTTCAGGAGCACCACTTCATGTGGGTGATCTACGAGTTG GACACTACCAAGGGTGACCATCAAACTCTCGATATGTTCTTCGAACCTGGCTACGTCCAAGAATCCCTGAAGCTG CAGTAAGAGTCATAACCTTGTTCGTTGCTTCTCGGTACCTGAGCTTGCCCAACTCTTTATAGTGCCGAAGGCAAA GGTCTCCATCGTACCGGTATCACCTCCTTCGCCTTCTTGCCCTTATCGAGCAGAGACCCGAAGGCCGCGGCTGCC TTGATTGATGAGCTCGAAGCTGACCTCAAGAAGAAGAAGGAGAGCGGAGAGATCTCGCCTGCTCTTGCCAAGCAA TATGACATCCAACTGGAGATTCTTCGCAATGACAAGGTTCCTGACATGGAATTCATTGTGTTCCCTGGTTATTTC ATGTGTCCCGGTATGTTGTCTCCGTCGCTATCTGGTGGATCATCGTTCCCTTACGGTTTCGATAGCGAGCCAGGG TACTGGTCGACGATTCATGACCGTCATCTCTTCTTTGAACCATCCAATCTCGAGAGGGTACATTGTGAGTCTTTG TCGGTTTCCTATACCAGTGATGTTCAGAGTTTCTAATTCGAAACAATAGCACATCAAGAGCAACAACCCCCTCGA TGATCCTGAAATTGACGCCAACTACTTTGGTCACAACTTTGGTGCGTTCTCATCCATTTGATGTTGCATAGATCT ATAGCTTGTACTTATCACGTCTCTGTCTACGCTCTGGCAGATCTCAAAGTTCTTTTGGAACACGTCAAGTACATT CGCTCTCTGGAAGATACCGAACCTTTCAAGTCGCACACCATCAAGGAGGTTGTTCCCGGTCCCGAATGCGCCAGC GACGCTGATATCGAGAGTACGTCATTCTTTTCTATGCATTCCTGGTGTGTCTGGTAATTGATTCATTGCCTGGGT TCTAGACCACATCAAGAATTCGCATATTACCACTTTCCGTAAGTCCTGTCTCATTCATTCCCCTCGTGTTGTCTA TGAACCCTACTAATCGTGTTGTTTCCTTGGCGCAGACACCGTTGGAAGCGCTTCGATGTTGCCTCGAGAGGATGG TGGTGTCGTTGACCCCAAGCTCAAGGTCTATGGTACCAAGAACCTCCGCGTTGCTGACCTTTCCATCGTTCCCCT TCACATCGCTGCTCACATGCAAGGTCAGTACTTCATACTGGGCGGTATGTCATTACCTCATTGCTAACCAACGTT CCTTTCGCGCTCCAGCCACTGCCTATGTGATCGGAGAGAAAGGTATGGCAGCAGCTGTGTGCATGCGTTGTTTCC TGTTCGCTGACGAGAACTTCTTTCTACTAGCTGCTGATTTGATCAAGGCGGATGCGAAGTAAAAGGCGAATGGAT ATATCGAAAAGAACAGGCAGTGTGTGTACATTATCTAGGGGTCGTTCTTTTCGGAACTGCAAGATATTTTTCGAT TT |
| Length | 2852 |