CopciAB_371647
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_371647 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | pgxC | Synonyms | 371647 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 28 family | |
| Location | scaffold_10:1508955..1511201 | Strand | + |
| Gene length (nt) | 2247 | Transcript length (nt) | 1525 |
| CDS length (nt) | 1368 | Protein length (aa) | 455 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262631 | 71.7 | 6.351E-216 | 668 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1437617 | 66.9 | 9.532E-202 | 627 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB19692 | 66.2 | 2.944E-199 | 620 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_121792 | 65.2 | 5.836E-195 | 607 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_131529 | 64.8 | 3.267E-193 | 602 |
| Auricularia subglabra | Aurde3_1_1314644 | 61.9 | 4.323E-180 | 565 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1048073 | 61.8 | 2.287E-180 | 565 |
| Pleurotus ostreatus PC9 | PleosPC9_1_59334 | 61.8 | 2.239E-180 | 565 |
| Flammulina velutipes | Flave_chr04AA00471 | 57.7 | 1.815E-165 | 522 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_10646 | 36.9 | 2.684E-89 | 301 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_38626 | 38.4 | 4.464E-63 | 224 |
| Grifola frondosa | Grifr_OBZ68558 | 44.2 | 1.299E-52 | 193 |
| Lentinula edodes B17 | Lened_B_1_1_11129 | 47.7 | 5.898E-27 | 115 |
| Lentinula edodes NBRC 111202 | Lenedo1_731020 | 24.3 | 2.332E-17 | 85 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | pgxC |
|---|---|
| Protein id | CopciAB_371647.T0 |
| Description | Belongs to the glycosyl hydrolase 28 family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00295 | Glycosyl hydrolases family 28 | IPR000743 | 130 | 372 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 27 | 0.9877 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR011050 | Pectin lyase fold/virulence factor |
| IPR000743 | Glycoside hydrolase, family 28 |
| IPR012334 | Pectin lyase fold |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004650 | polygalacturonase activity | MF |
| GO:0005975 | carbohydrate metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| K01213 |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 28 family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH28 |
Transcription factor
| Group |
|---|
| No records |
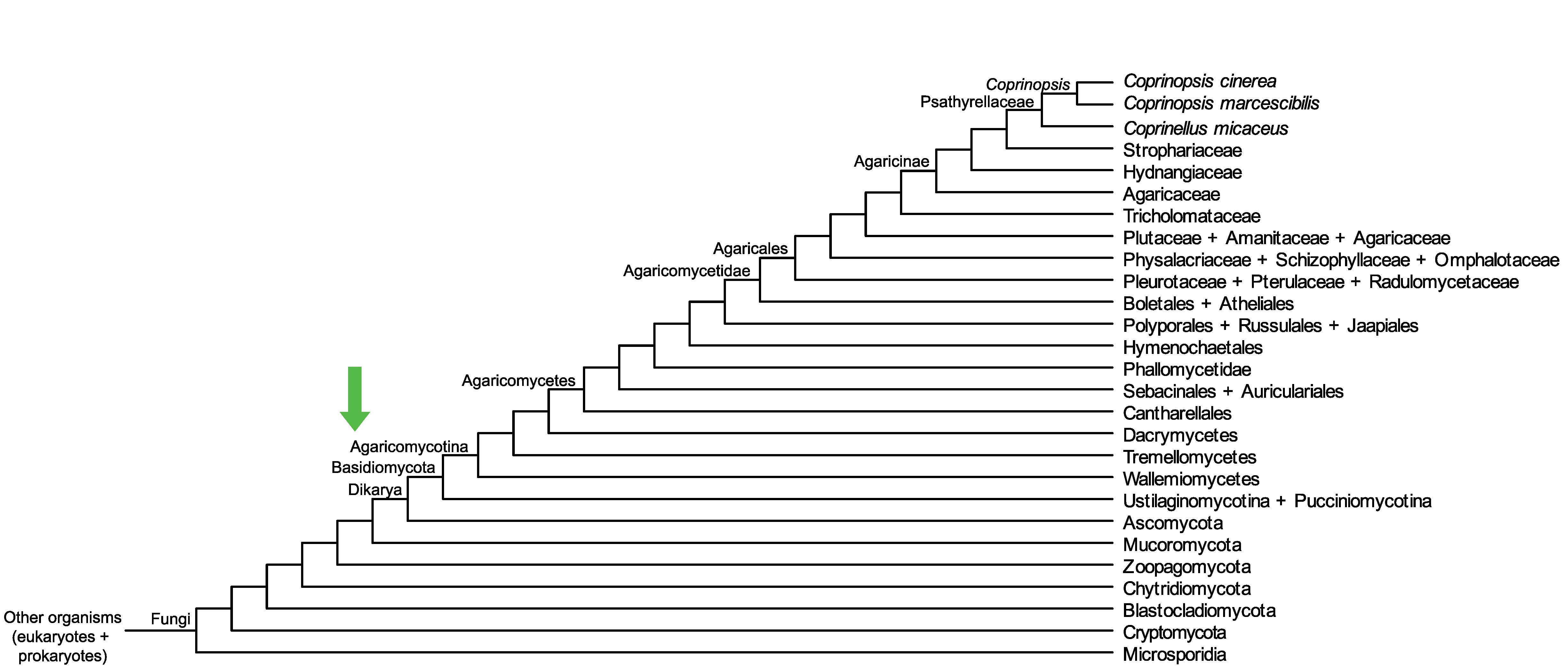
Conservation of CopciAB_371647 across fungi.
Arrow shows the origin of gene family containing CopciAB_371647.
Protein
| Sequence id | CopciAB_371647.T0 |
|---|---|
| Sequence |
>CopciAB_371647.T0 MGKTSLMLRFIASSLWLCLLFAALGRAQDLDADEVLEAGAPPRRCIIPYSRNPNQDDSQSILRTVNRCSTNSIIE FRPANYSVYTPISLLNLRNVKILLNGNIVLPSNITKVQVEINNTLNQPALYATPWIYIQGTNVELVGSSDFKYGR FYSFGQQWWDLGIRTLRPHLATFNVTNGLLKNLKIIKPIAWGWSIPGQNIRIENHFVDAKPENATRDDTVSFPFN TDGFNLSGKNITIDGYYGHNGDDCVSVINGARDIVAKNGYCGFSSHGLSIGSLGRDGAEHTVKNVLFKNWTMDGA VYGARFKSWTGGRGYGENIAWEDITLVNVSTGIFITQNYYDQDKGPRPPNPGNTSTKVINISFRNFKGSLGTNWT DGSCISTPCWNYVEGLDEPKAVILDLYPDTATNVTLENIDIHPFNQPGEVANVMCDPTALSAREQSRLGFKCTNG PFEAT |
| Length | 455 |
Coding
| Sequence id | CopciAB_371647.T0 |
|---|---|
| Sequence |
>CopciAB_371647.T0 ATGGGGAAGACGAGCTTGATGTTGAGGTTTATTGCTTCGTCGCTTTGGCTCTGCCTCCTCTTTGCTGCTCTTGGG CGTGCCCAGGATCTCGACGCCGACGAGGTACTGGAGGCGGGGGCACCCCCTCGTAGATGCATTATTCCGTACTCC AGGAATCCGAACCAGGACGACTCGCAGTCTATCCTGCGCACTGTGAATAGGTGCTCGACCAACTCGATCATCGAG TTTAGGCCGGCCAATTATAGTGTTTACACTCCCATCTCGCTTTTGAATCTTCGAAACGTCAAGATTCTTTTGAAT GGGAATATTGTCCTCCCTTCGAATATAACGAAGGTGCAAGTCGAGATTAACAATACGCTGAACCAGCCTGCTCTT TATGCCACGCCCTGGATCTACATCCAAGGGACGAACGTCGAGCTCGTTGGATCAAGCGATTTCAAGTACGGGCGC TTCTACAGTTTCGGGCAGCAATGGTGGGACTTGGGTATCCGGACCCTCCGACCGCACCTTGCAACATTTAACGTC ACGAACGGCCTTTTGAAGAACCTCAAGATTATCAAACCCATTGCTTGGGGATGGAGTATCCCCGGCCAGAACATC AGGATTGAAAACCATTTCGTGGATGCGAAGCCCGAGAATGCGACGCGGGATGATACAGTCAGCTTCCCTTTCAAT ACCGATGGGTTCAACTTGTCTGGCAAGAATATCACCATTGACGGGTACTACGGACACAATGGCGACGATTGCGTT TCTGTCATCAACGGAGCTCGAGACATTGTCGCGAAGAACGGGTACTGTGGATTTTCCTCCCATGGACTCTCCATC GGCTCTTTGGGGCGTGATGGGGCGGAGCATACAGTTAAAAATGTGTTATTCAAGAATTGGACTATGGATGGAGCT GTGTATGGCGCTCGGTTCAAGAGTTGGACTGGGGGAAGGGGATATGGAGAGAACATTGCTTGGGAGGATATCACG CTCGTCAACGTATCCACCGGTATATTCATCACTCAGAACTACTATGACCAAGACAAAGGACCGCGCCCACCCAAT CCAGGCAACACAAGCACCAAAGTGATCAATATATCCTTCAGGAACTTTAAAGGAAGTCTAGGTACCAATTGGACG GACGGATCGTGTATAAGCACGCCTTGTTGGAATTACGTGGAGGGGCTTGATGAACCTAAAGCTGTTATTTTGGAT TTGTATCCTGATACTGCGACGAATGTCACGTTGGAGAACATCGATATACACCCATTCAACCAGCCTGGAGAGGTG GCGAATGTGATGTGCGACCCGACGGCTCTAAGCGCGAGGGAGCAGAGTAGGCTGGGATTCAAGTGCACCAACGGA CCTTTCGAGGCGACGTAG |
| Length | 1368 |
Transcript
| Sequence id | CopciAB_371647.T0 |
|---|---|
| Sequence |
>CopciAB_371647.T0 AGGGGTTGGACTACATTCGACCTCATTCCATCTTTCACCGGCGCTTTGCAGAAGGAGGCATGGGGAAGACGAGCT TGATGTTGAGGTTTATTGCTTCGTCGCTTTGGCTCTGCCTCCTCTTTGCTGCTCTTGGGCGTGCCCAGGATCTCG ACGCCGACGAGGTACTGGAGGCGGGGGCACCCCCTCGTAGATGCATTATTCCGTACTCCAGGAATCCGAACCAGG ACGACTCGCAGTCTATCCTGCGCACTGTGAATAGGTGCTCGACCAACTCGATCATCGAGTTTAGGCCGGCCAATT ATAGTGTTTACACTCCCATCTCGCTTTTGAATCTTCGAAACGTCAAGATTCTTTTGAATGGGAATATTGTCCTCC CTTCGAATATAACGAAGGTGCAAGTCGAGATTAACAATACGCTGAACCAGCCTGCTCTTTATGCCACGCCCTGGA TCTACATCCAAGGGACGAACGTCGAGCTCGTTGGATCAAGCGATTTCAAGTACGGGCGCTTCTACAGTTTCGGGC AGCAATGGTGGGACTTGGGTATCCGGACCCTCCGACCGCACCTTGCAACATTTAACGTCACGAACGGCCTTTTGA AGAACCTCAAGATTATCAAACCCATTGCTTGGGGATGGAGTATCCCCGGCCAGAACATCAGGATTGAAAACCATT TCGTGGATGCGAAGCCCGAGAATGCGACGCGGGATGATACAGTCAGCTTCCCTTTCAATACCGATGGGTTCAACT TGTCTGGCAAGAATATCACCATTGACGGGTACTACGGACACAATGGCGACGATTGCGTTTCTGTCATCAACGGAG CTCGAGACATTGTCGCGAAGAACGGGTACTGTGGATTTTCCTCCCATGGACTCTCCATCGGCTCTTTGGGGCGTG ATGGGGCGGAGCATACAGTTAAAAATGTGTTATTCAAGAATTGGACTATGGATGGAGCTGTGTATGGCGCTCGGT TCAAGAGTTGGACTGGGGGAAGGGGATATGGAGAGAACATTGCTTGGGAGGATATCACGCTCGTCAACGTATCCA CCGGTATATTCATCACTCAGAACTACTATGACCAAGACAAAGGACCGCGCCCACCCAATCCAGGCAACACAAGCA CCAAAGTGATCAATATATCCTTCAGGAACTTTAAAGGAAGTCTAGGTACCAATTGGACGGACGGATCGTGTATAA GCACGCCTTGTTGGAATTACGTGGAGGGGCTTGATGAACCTAAAGCTGTTATTTTGGATTTGTATCCTGATACTG CGACGAATGTCACGTTGGAGAACATCGATATACACCCATTCAACCAGCCTGGAGAGGTGGCGAATGTGATGTGCG ACCCGACGGCTCTAAGCGCGAGGGAGCAGAGTAGGCTGGGATTCAAGTGCACCAACGGACCTTTCGAGGCGACGT AGCTTCTTCAGCGTGGCTACCATCTCTTTCAGTCGACTACCTTATAGGTGTCATGTGTGTACTTGTCTGCATTGA ATCAAACTTTAGTCTTCAAGGTGGA |
| Length | 1525 |
Gene
| Sequence id | CopciAB_371647.T0 |
|---|---|
| Sequence |
>CopciAB_371647.T0 AGGGGTTGGACTACATTCGACCTCATTCCATCTTTCACCGGCGCTTTGCAGAAGGAGGCATGGGGAAGACGAGCT TGATGTTGAGGTTTATTGCTTCGTCGCTTTGGCTCTGCCTCCTCTTTGCTGCTCTTGGGCGTGCCCAGGATCTCG ACGCCGACGAGGTACTGGAGGCGGGGGCACCCCCTCGTAGATGCATTATTCCGTACTCCAGGAATCCGAACCAGG ACGACTCGCAGTCTATCCTGCGCACTGTGAATAGGTGCTCGACCAACTCGATCATCGAGTTTAGGCCGGCCAATT ATAGTGTTTACACTCCCATCTCGCTTTTGAATCTTCGTAAGTGCTTCATTTAAAGTTTGGGACTGGGTGCGGCGA CTGATGGTATGTACGTTGCATCTGGTGGTTAGGAAACGTCAAGATTCTTTTGAATGGGAATATTGTCCTCCCTTC GAATATAACGAAGGTGCAAGTCGAGATTAACAATACGCTGAACCAGCCTGCTGTGCGTGTTTGCGACTTTTTCGA GGTCACCTTTGTGCTGATGGTATAGCTGTAGCTTTATGCCACGCCCTGGATCTACATCCAAGGGACGAACGTCGA GCTCGTTGGATCAAGCGATTTCAAGTACGGGCGCTTCTACAGTTTCGGGCAGCAATGGTGGGACTTGGGTATCCG GGTACGCCTTTCTTCCGTGACTCTACCTCTCCCTCTATCCATTATTGACGCCTGTGAATTAGACCCTCCGACCGC ACCTTGCAACATTTAACGTCACGAACGGCCTTTTGAAGAACCTCAAGATTATCAAACCCATTGCTTGGGGATGGA GTATCCCCGGCCAGAACATCAGGATTGAAAACCATTTCGTGGATGCGAAGCCCGAGAATGCGACGCGGGATGATA CAGTCGTGAGTCTCCCTCTCTTTTAACGTGTGGCCCCAAAGTATGTGCTCTAATGCATGCGTTTGGTTTTTTGTA GAGCTTCCCTTTCAATACGTACGTGGATGGTGTTTTTCCAGTGCGAGTGATTGGATGTTTATTGTCGCTAGCGAT GGGTTCAACTTGTCTGGCAAGAATATCACCATTGACGGGTGTGTTTCCTCTTTCCAGCGCCTTAATTCCAGAGGT TGTCATTTCCTAACCGTCGGATATCAGGTACTACGGACACAATGGCGACGATTGCGTTTCTGTCATCAACGGAGC TCGAGACATTGTCGCGAAGAACGGGTACTGTGGATTTTCCTCCCATGGACTCTCCATCGGCTCTTTGGGGCGTGA TGGGGCGGAGCATACAGTTAAAAATGTGTTATTCAAGAATTGGACTATGGATGGAGCTGTGTATGGCGCTCGGGT AAGTTGAACCCTCATTCGTGGTGCGGCCCGTGATGCTGACGCTAATGCTGCACACAGTTCAAGAGTTGGACTGGG GGAAGGGGATATGGAGAGAACATTGCTTGGGAGGATATCACGCTCGTCAACGTATCCACCGGTATATTCATCACT CAGAAGTGAGTATCGTTCCTAGCATCGCTGGTACCTGAGTGACATCGCTAACCTGTATGAATCCGATCAGCTACT ATGACCAAGTGAGTCACTTTAAATTCCTCAGCACCCCCATTTCTAACGACGTCGCTATAGGACAAAGGACCGCGC CCACCCAATCCAGGCAACACAAGCACCAAAGTGATCAATATATCCTTCAGGAACTTTAAAGGAAGTCTAGGTACC AATTGGAGTAAGTCATTCGCCAAATCGGGGTACGGTCTTGGCTTATTGACGGTCCTGTCACCGATCGCGCCCCTA TTTTCGTTTTTGAACCTGCTTTGAAAATGCGAAACTACGTGAAACTATGTTAGCGGACGGATCGTGTATAAGCAC GCCTTGTTGGAATTACGTGGAGGGGCTTGATGAACCTAAAGCTGTTATTTTGGATTTGTATCCTGATACTGGTGA GTCAGGTTCGGTGGATGCGGGGTGCGGTGGCTGATCGTGTTGTACAGCGACGAATGTCACGTTGGAGAACATCGA TATACACCCATTCAACCAGCCTGGAGAGGTGGCGAATGTGATGTGCGACCCGACGGCTCTAAGCGCGAGGGAGCA GAGTAGGCTGGGATTCAAGTGCACCAACGGACCTTTCGAGGCGACGTAGCTTCTTCAGCGTGGCTACCATCTCTT TCAGTCGACTACCTTATAGGTGTCATGTGTGTACTTGTCTGCATTGAATCAAACTTTAGTCTTCAAGGTGGA |
| Length | 2247 |