CopciAB_373700
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_373700 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 373700 |
| Uniprot id | Functional description | Cellulase (glycosyl hydrolase family 5) | |
| Location | scaffold_7:465822..469250 | Strand | + |
| Gene length (nt) | 3429 | Transcript length (nt) | 2198 |
| CDS length (nt) | 1866 | Protein length (aa) | 621 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7262821 | 72.1 | 1.218E-304 | 974 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_114151 | 70.4 | 6.298E-291 | 934 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_14671 | 69.1 | 1.964E-291 | 930 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14918 | 66.5 | 3.616E-291 | 915 |
| Lentinula edodes NBRC 111202 | Lenedo1_1000518 | 70.1 | 1.047E-283 | 873 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87373 | 68.1 | 1.457E-268 | 829 |
| Pleurotus ostreatus PC15 | PleosPC15_2_45975 | 67.9 | 7.319E-268 | 827 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_72051 | 67.8 | 1.402E-267 | 826 |
| Flammulina velutipes | Flave_chr10AA00936 | 59.4 | 6.34E-252 | 781 |
| Schizophyllum commune H4-8 | Schco3_81800 | 62.9 | 7.794E-252 | 781 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1568598 | 64.3 | 1.534E-251 | 780 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_373700.T0 |
| Description | Cellulase (glycosyl hydrolase family 5) |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 172 | 323 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| 1 | 13 | 35 | 22 |
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR001547 | Glycoside hydrolase, family 5 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| S | Cellulase (glycosyl hydrolase family 5) |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_9 |
Transcription factor
| Group |
|---|
| No records |
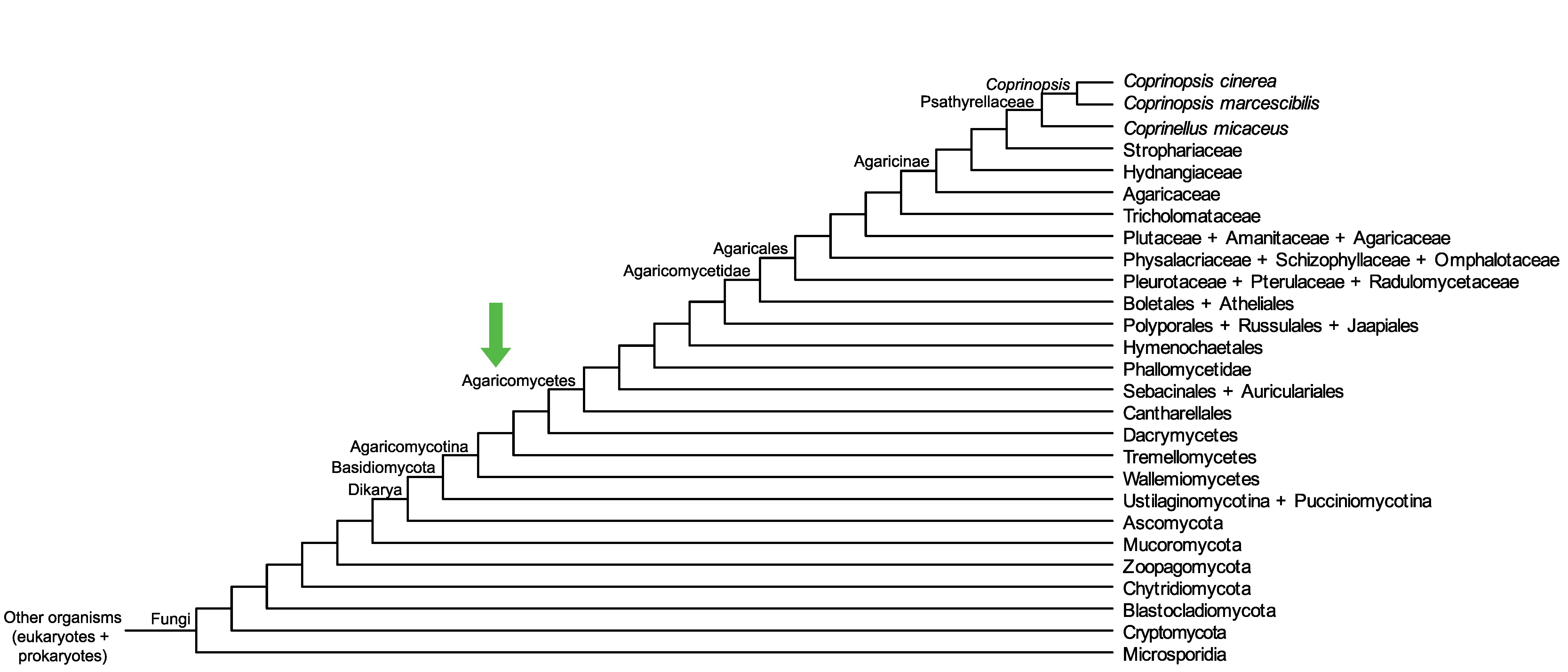
Conservation of CopciAB_373700 across fungi.
Arrow shows the origin of gene family containing CopciAB_373700.
Protein
| Sequence id | CopciAB_373700.T0 |
|---|---|
| Sequence |
>CopciAB_373700.T0 MGPGKPMKKGVKISIAVAVLIAVITIAIAVPLAVTVGKSKNGTSSSGGGAGGGSSGGGSATSGKSGSLVTMEDGS TFTYVNEFGGDWAADPARPFGSGGKAQEWSPRVGVEEWKWGEHVIRGVNLGGWLVTEPFICPELYERFIENDENV TVVDEWTLSLAMGDRLPEEMENHYKTFITEQDFAEIAAAGLNWIRVPIGYWAIETMGEEPFLVGTSWTYFLKAIQ WARKYGLRIYLDLHALPGSQNGWNHSGKAGTVNFMHGTMGIANAQRTLTYLRIFTQFVSQPQYKDVVPIVGIVNE ILWDTIGEEAVQSFYYAAYEAMRGASGIGEGNGPYIAIHEGFQGPAIWEGFLLGSDRVVLDQHPYLAFMGDPNST PESSAPRPCEWAIATNQSQQVFGVTVGGEFSTAINNCGLWLNGVDGAPIPGCELWDDWNSWNSTIISGLKQVTLA SMDALQHFFFWTWKIGNSTRLGTSSSPMWHYKLGLERGWIPKDPREAIGHCAEVLGTSQLFDGRYPSTATGGAAA GTLDPTRASAHPFPPATISPSFSGDLLSLLPTYTPTGTLQTLFAPTFTAAPSAVVGTGWNNPDDNSPAYVPVAGC SYPDAWDAVDVPLSEPFCTGS |
| Length | 621 |
Coding
| Sequence id | CopciAB_373700.T0 |
|---|---|
| Sequence |
>CopciAB_373700.T0 ATGGGTCCAGGCAAACCGATGAAAAAAGGCGTCAAGATCAGTATCGCGGTGGCCGTTCTTATTGCTGTCATTACG ATTGCGATAGCCGTCCCGTTGGCCGTCACGGTTGGGAAGAGCAAGAATGGCACGAGTTCGTCGGGGGGAGGGGCT GGGGGTGGCTCCAGTGGGGGTGGGTCGGCGACGTCTGGAAAGTCTGGGAGTCTCGTTACGATGGAGGACGGTTCG ACGTTCACGTATGTGAACGAGTTTGGGGGCGACTGGGCTGCTGACCCTGCGCGTCCCTTTGGTTCGGGTGGAAAG GCCCAGGAGTGGTCGCCACGCGTCGGTGTGGAAGAATGGAAATGGGGAGAACATGTTATTCGTGGCGTCAACCTG GGAGGGTGGTTAGTCACCGAACCGTTCATCTGCCCCGAGTTATATGAGAGATTTATCGAGAATGATGAAAATGTG ACCGTGGTCGACGAGTGGACCCTTTCGCTAGCTATGGGCGACCGCCTTCCAGAAGAGATGGAAAACCATTACAAG ACCTTCATCACTGAACAAGACTTTGCAGAAATTGCAGCCGCCGGACTGAACTGGATAAGAGTTCCCATCGGTTAT TGGGCAATTGAGACGATGGGCGAGGAGCCTTTCCTCGTAGGTACATCGTGGACCTATTTCCTGAAGGCGATTCAA TGGGCGAGAAAATACGGTCTTCGAATTTACCTCGATCTCCATGCTCTCCCGGGAAGCCAAAATGGCTGGAACCAT TCTGGAAAAGCTGGCACTGTTAATTTTATGCACGGCACAATGGGAATCGCCAATGCTCAAAGAACACTTACGTAT CTGCGAATTTTCACGCAATTTGTGAGCCAACCTCAGTACAAGGACGTTGTTCCGATTGTTGGTATAGTGAACGAA ATTCTTTGGGATACTATTGGCGAGGAAGCGGTTCAGAGCTTCTATTACGCCGCCTATGAGGCCATGAGGGGTGCT TCAGGTATTGGTGAAGGAAATGGCCCCTATATTGCTATCCATGAGGGCTTCCAAGGGCCTGCGATCTGGGAAGGA TTCCTTTTAGGCTCGGATCGAGTAGTTCTTGACCAGCACCCGTACTTGGCGTTCATGGGTGATCCAAACAGCACC CCTGAATCTAGTGCTCCCAGACCCTGTGAATGGGCGATCGCCACCAACCAGTCCCAGCAAGTTTTCGGCGTGACG GTTGGCGGAGAATTCTCGACTGCTATTAACAACTGTGGACTCTGGTTGAACGGCGTCGACGGAGCTCCTATTCCT GGATGCGAACTTTGGGATGATTGGAACTCGTGGAACTCGACGATAATCTCGGGATTGAAACAAGTCACGCTGGCT TCTATGGATGCCTTGCAGCACTTTTTCTTCTGGACTTGGAAGATAGGTAATTCTACGAGGCTTGGCACGTCCTCC TCGCCGATGTGGCACTATAAACTTGGTTTGGAAAGGGGCTGGATCCCGAAAGATCCGAGGGAAGCTATTGGACAC TGTGCTGAAGTTTTGGGCACGTCTCAATTATTCGATGGTCGATACCCGTCGACTGCGACCGGAGGCGCTGCTGCT GGGACACTTGACCCTACCCGAGCTTCAGCGCACCCCTTCCCGCCAGCTACGATTTCACCCTCTTTCTCGGGTGAT TTACTCTCTCTTCTGCCCACCTATACTCCGACAGGGACGCTGCAGACATTGTTTGCCCCTACATTCACCGCTGCT CCGTCTGCTGTGGTGGGCACCGGTTGGAATAATCCCGATGATAATTCGCCCGCCTATGTTCCCGTAGCTGGTTGT TCATATCCTGATGCGTGGGATGCTGTCGATGTGCCGCTCTCAGAACCGTTCTGCACCGGCTCTTGA |
| Length | 1866 |
Transcript
| Sequence id | CopciAB_373700.T0 |
|---|---|
| Sequence |
>CopciAB_373700.T0 ACCACGTTCCCTACTCTTTCTCGCCACCAGACAGCGGTGACCGTGCCCTCCCTGAAGAGTCGACGACGAGTATGG GTCCAGGCAAACCGATGAAAAAAGGCGTCAAGATCAGTATCGCGGTGGCCGTTCTTATTGCTGTCATTACGATTG CGATAGCCGTCCCGTTGGCCGTCACGGTTGGGAAGAGCAAGAATGGCACGAGTTCGTCGGGGGGAGGGGCTGGGG GTGGCTCCAGTGGGGGTGGGTCGGCGACGTCTGGAAAGTCTGGGAGTCTCGTTACGATGGAGGACGGTTCGACGT TCACGTATGTGAACGAGTTTGGGGGCGACTGGGCTGCTGACCCTGCGCGTCCCTTTGGTTCGGGTGGAAAGGCCC AGGAGTGGTCGCCACGCGTCGGTGTGGAAGAATGGAAATGGGGAGAACATGTTATTCGTGGCGTCAACCTGGGAG GGTGGTTAGTCACCGAACCGTTCATCTGCCCCGAGTTATATGAGAGATTTATCGAGAATGATGAAAATGTGACCG TGGTCGACGAGTGGACCCTTTCGCTAGCTATGGGCGACCGCCTTCCAGAAGAGATGGAAAACCATTACAAGACCT TCATCACTGAACAAGACTTTGCAGAAATTGCAGCCGCCGGACTGAACTGGATAAGAGTTCCCATCGGTTATTGGG CAATTGAGACGATGGGCGAGGAGCCTTTCCTCGTAGGTACATCGTGGACCTATTTCCTGAAGGCGATTCAATGGG CGAGAAAATACGGTCTTCGAATTTACCTCGATCTCCATGCTCTCCCGGGAAGCCAAAATGGCTGGAACCATTCTG GAAAAGCTGGCACTGTTAATTTTATGCACGGCACAATGGGAATCGCCAATGCTCAAAGAACACTTACGTATCTGC GAATTTTCACGCAATTTGTGAGCCAACCTCAGTACAAGGACGTTGTTCCGATTGTTGGTATAGTGAACGAAATTC TTTGGGATACTATTGGCGAGGAAGCGGTTCAGAGCTTCTATTACGCCGCCTATGAGGCCATGAGGGGTGCTTCAG GTATTGGTGAAGGAAATGGCCCCTATATTGCTATCCATGAGGGCTTCCAAGGGCCTGCGATCTGGGAAGGATTCC TTTTAGGCTCGGATCGAGTAGTTCTTGACCAGCACCCGTACTTGGCGTTCATGGGTGATCCAAACAGCACCCCTG AATCTAGTGCTCCCAGACCCTGTGAATGGGCGATCGCCACCAACCAGTCCCAGCAAGTTTTCGGCGTGACGGTTG GCGGAGAATTCTCGACTGCTATTAACAACTGTGGACTCTGGTTGAACGGCGTCGACGGAGCTCCTATTCCTGGAT GCGAACTTTGGGATGATTGGAACTCGTGGAACTCGACGATAATCTCGGGATTGAAACAAGTCACGCTGGCTTCTA TGGATGCCTTGCAGCACTTTTTCTTCTGGACTTGGAAGATAGGTAATTCTACGAGGCTTGGCACGTCCTCCTCGC CGATGTGGCACTATAAACTTGGTTTGGAAAGGGGCTGGATCCCGAAAGATCCGAGGGAAGCTATTGGACACTGTG CTGAAGTTTTGGGCACGTCTCAATTATTCGATGGTCGATACCCGTCGACTGCGACCGGAGGCGCTGCTGCTGGGA CACTTGACCCTACCCGAGCTTCAGCGCACCCCTTCCCGCCAGCTACGATTTCACCCTCTTTCTCGGGTGATTTAC TCTCTCTTCTGCCCACCTATACTCCGACAGGGACGCTGCAGACATTGTTTGCCCCTACATTCACCGCTGCTCCGT CTGCTGTGGTGGGCACCGGTTGGAATAATCCCGATGATAATTCGCCCGCCTATGTTCCCGTAGCTGGTTGTTCAT ATCCTGATGCGTGGGATGCTGTCGATGTGCCGCTCTCAGAACCGTTCTGCACCGGCTCTTGAATTCATTCATTCA TTTTATTTTTTCATGCTTTCGCATGGATAGTTATGGCTGTGGCATGACCCCCCGAGCATGACATGATGACCCTGT GTAGTGTACGCTGCTTTCTTGGACGATGTTGTGACCTGACTGTTTGCTTCATCTTCTTTTGTTCTCGCATTTCTT GGACTACTTATTGGAAGTGATATGGACTTTTTGCATCTCTGTATTTTATGTAGATGAGCGTAGATGCTAATGCTA TGTGATGCCATGATTTCTTTTGA |
| Length | 2198 |
Gene
| Sequence id | CopciAB_373700.T0 |
|---|---|
| Sequence |
>CopciAB_373700.T0 ACCACGTTCCCTACTCTTTCTCGCCACCAGACAGCGGTGACCGTGCCCTCCCTGAAGAGTCGACGACGAGTATGG GTCCAGGCAAACCGATGAAAAAAGGCGTCAAGATCAGTATCGCGGTGGCCGTTCTTATTGCTGTCATTACGATTG CGATAGCCGTCCCGTTGGCCGTCACGGTTGGGAAGAGCAAGAATGGCACGAGTTCGTCGGGGGGAGGGGCTGGGG GTGGCTCCAGTGGGGGTGGGTCGGCGACGTCTGGAAAGTCTGGGAGTCTCGTTACGATGGAGGACGGTTCGACGT TCACGTATGTGAACGAGTTTGGGGGCGACTGGGCTGCTGACCCTGCGCGTCCCTTTGGTTCGGGTGGAAAGGCCC AGGAGTGGTCGCCACGCGTCGGTGTGGAAGAATGGAAATGGGGAGAACATGTTATTCGTGGCGTCAACCTGGGGT ACGTTTTCTTTAACCGTATAGACGCGTACTTTGCTCACTTGGCGACTATTTCTTCCTAAAGAGGGTGGTTAGGTG AGTATGGTCTTTGTGGTCTGTTTCCACGTTCTGATGTTTGCAGTCACCGAACCGTTCATCTGCCCCGAGTTATAT GAGAGATTTATCGAGAATGATGAAAATGTGACCGTGGTCGACGAGTGGACCCTTTCGCTAGCTATGGGCGACCGC CTTCCAGAAGAGATGGAAAACCATTACAAGACCTTCATCGTGTGTTGTCCTACTGGCTTCTATTCTCGTCAGCGA CTGATCACGTCCAACAGACTGAACAAGACTTTGCAGAAATTGCAGGTATGAGCGGACAAGCCTCTTCACAACTGG TCGAATGTCAACATGTCTATTTTAGCCGCCGGACTGAACTGGATAAGAGTTCCCATCGGTTATTGGGCAATTGAG ACGATGGGCGAGGAGCCTTTCCTCGTAGGTACATCGTGGACCTATTTCCTGAAGGCGTGAGTGCTCTGATACCGC ATTCGAATCCCTTTGACGCTGATGATACCGCGTAGGATTCAATGGGCGAGAAAATACGGTCTTCGAATTTACCTC GATCTCCATGCTCTCCCGGGAAGCCAAAATGGCTGGGTAAGTATCTCATCTTGTTCACAAAAACTTCCCGCTCAA TGATGTCTATGATAGAACCATTCTGGAAAAGGTTTGCTTGCCTCTACACCGCCATTCGTCGAAAATAGACTGACA AATGCCGTTCTCACTGTGAAGCTGGCACTGTTAATTTGTGAGATCATTGTGACATCCGTTCCGTTGGTTCTGAAT CTGGTGCTAATCGCGAAATTCTTTTCCAGTATGCACGGCACAATGGGAATCGCCAATGCTCAAAGAACACTTACG TATCTGCGAATTTTCACGCAATTTGTGAGCCAACCTCAGTACAAGGACGTTGTTCCGATGTGAGTGAGAAGTATC GGTGGTTTCTCGCCTTGACTGAACACCCTTCTTTCAAGTGTTGGTATAGTGAACGAAATTCTTTGGGATACTATT GGCGAGGAAGCGGTTCAGAGCTTCTATTACGCCGCCTATGAGGCCATGAGGGGTGCTTCGTACGTTACTCGAACT TCACCTCGGTCGACTGAACGTTAACGTATCTATTGCAGAGGTATTGGTGAAGGAAATGGCCCCTATATTGCTATC CATGAGGGCTTCCAAGGGGTACGTCCGGGATTTCTAGGTTCAGTTCATTATCTGAATGATGTTAACTTGGTTTGC GTAGCCTGCGATCTGGGAAGGGTACGCCTTCTCCCACTGGTTCTGGTTCTGGTGTACTCAATGAATTTTAGATTC CTTTTAGGCTCGGATCGAGTAGTTCTTGACCAGCACCCGGTACGTTCGTTCCTTTTCTCTAGATGCCTTGTACTC AACGGAGTCGATAGTACTTGGCGTTCATGGGTGATCCAAACAGCACGTAAGTCTCGTCTTGTGGAGTTCTTTGAC CAGAGCTAGACCCTTTCTTTCGATAGCCCTGAATCTAGTGCTCCCAGACCCTGGTAAGGACGATTTTGTCGGTTG ATGTTAAAGTTATTCATTGGATCTGCAGTGAATGGGCGATCGCCACCAACCAGTCCCAGCAAGTTTTCGGCGTGA CGGTTGGCGGAGAATTCTCGACTGCTATTAACAACTGTGGACTCTGGTTCGTTCTTGGTACCCTCTCCAGGCTTC TCGACTCACACGAGATCGTCCTTAGGTTGAACGGCGTCGACGGAGCTCCTATTCCTGGATGCGAACTTTGGGATG ATTGGAACGTAAGTCTTCCATCCTTCTTCCTTCCTTTCCGCGATTCCCGATTGTTAAGTTGTCCTTCGTATCATC CAGTCGTGGAACTCGACGATAATCTCGGGATTGAAACAAGTCACGCTGGCTTCTATGGATGCCTTGCAGCACTTT TTCTTCTGGACTTGGAAGATAGGTAATTCTACGAGGCTTGGCACGTCCTCCTCGCCGATGTGGCACTATAAACTT GGTTTGGAAAGGGGCTGGATCCCGAAAGGTATGCCGCCCAGCTCGTTTTTGTTGGCTCGCTGGTTTTACTCACAA GGTGGGGTCCCCGTGTAGATCCGAGGGAAGCTATTGGACACTGTGCTGAAGTTTTGGGCACGTCTCAATTATTCG ATGGTCGATACCCGTCGACTGCGACCGGAGGCGTAAGTAATTTACCCACTGTTATGTCACTCTTGAGGCGCCCTT GTGCTGATTATGTTAACCCTTCTTTCGCAGGCTGCTGCTGGGACACTTGACCCTACCCGAGCTTCAGCGCACCCC TTCCCGCCAGCTACGATTTCACCCTCTTTCTCGGGTGATTTACTCTCTCTTCTGCCCACCTATACTCCGACAGGG ACGCTGCAGACATTGTTTGCCCCTACATTCACCGCTGCTCCGTCTGCTGTGGTGGGCACCGGTTGGAATAATCCC GATGATAATTCGCCCGCCTATGTGTAAGTTTCTGCCGCGTTTTTTTCTGCATTCCATTTTCCGTGAGACTCATTT ACTGTTCTGCCGTTTCATGATATAGTCCCGTAGCTGGTTGTTCATATCCTGAGTGGGTGCCTGAAACTTCTTGAC TTCTCTCTCTCAGATCACTAACGCTTGGTTCTTTGTAGTGCGTGGGATGCTGTCGATGTGCCGCTCTCAGAACCG TTCTGCACCGGCTCTTGAATTCATTCATTCATTTTATTTTTTCATGCTTTCGCATGGATAGTTATGGCTGTGGCA TGACCCCCCGAGCATGACATGATGACCCTGTGTAGTGTACGCTGCTTTCTTGGACGATGTTGTGACCTGACTGTT TGCTTCATCTTCTTTTGTTCTCGCATTTCTTGGACTACTTATTGGAAGTGATATGGACTTTTTGCATCTCTGTAT TTTATGTAGATGAGCGTAGATGCTAATGCTATGTGATGCCATGATTTCTTTTGA |
| Length | 3429 |