CopciAB_376354
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_376354 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | NAG1 | Synonyms | 376354 |
| Uniprot id | Functional description | Glycoside hydrolase family 20 protein | |
| Location | scaffold_7:859646..861914 | Strand | - |
| Gene length (nt) | 2269 | Transcript length (nt) | 1830 |
| CDS length (nt) | 1665 | Protein length (aa) | 554 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN1502_nagA |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB14630 | 71.8 | 1.177E-277 | 852 |
| Agrocybe aegerita | Agrae_CAA7261537 | 67.6 | 2.312E-259 | 799 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_120598 | 66.8 | 2.425E-255 | 787 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1526800 | 65 | 3.427E-245 | 758 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_186352 | 64.5 | 5.581E-245 | 757 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_116762 | 64.1 | 1.07E-239 | 742 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1094009 | 63 | 1.188E-237 | 736 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_13165 | 62.2 | 6.766E-237 | 734 |
| Lentinula edodes NBRC 111202 | Lenedo1_1052620 | 61.6 | 3.656E-235 | 729 |
| Flammulina velutipes | Flave_chr06AA00028 | 60.3 | 2.786E-228 | 709 |
| Grifola frondosa | Grifr_OBZ71292 | 61.8 | 3.489E-216 | 674 |
| Lentinula edodes B17 | Lened_B_1_1_3886 | 59.9 | 3.279E-215 | 671 |
| Auricularia subglabra | Aurde3_1_1302918 | 57 | 4.629E-210 | 657 |
| Pleurotus ostreatus PC9 | PleosPC9_1_87509 | 69 | 9.586E-174 | 551 |
| Schizophyllum commune H4-8 | Schco3_66483 | 59.6 | 8.44E-149 | 479 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | NAG1 |
|---|---|
| Protein id | CopciAB_376354.T0 |
| Description | Glycoside hydrolase family 20 protein |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd06562 | GH20_HexA_HexB-like | - | 177 | 532 |
| Pfam | PF14845 | beta-acetyl hexosaminidase like | IPR029019 | 20 | 154 |
| Pfam | PF00728 | Glycosyl hydrolase family 20, catalytic domain | IPR015883 | 177 | 509 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 19 | 0.9839 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR017853 | Glycoside hydrolase superfamily |
| IPR025705 | Beta-hexosaminidase |
| IPR015883 | Glycoside hydrolase family 20, catalytic domain |
| IPR029019 | Beta-hexosaminidase, eukaryotic type, N-terminal |
| IPR029018 | Beta-hexosaminidase-like, domain 2 |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004563 | beta-N-acetylhexosaminidase activity | MF |
| GO:0005975 | carbohydrate metabolic process | BP |
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
KEGG
| KEGG Orthology |
|---|
| K12373 |
EggNOG
| COG category | Description |
|---|---|
| G | Glycoside hydrolase family 20 protein |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH20 |
Transcription factor
| Group |
|---|
| No records |
Conservation of CopciAB_376354 across fungi.
Arrow shows the origin of gene family containing CopciAB_376354.
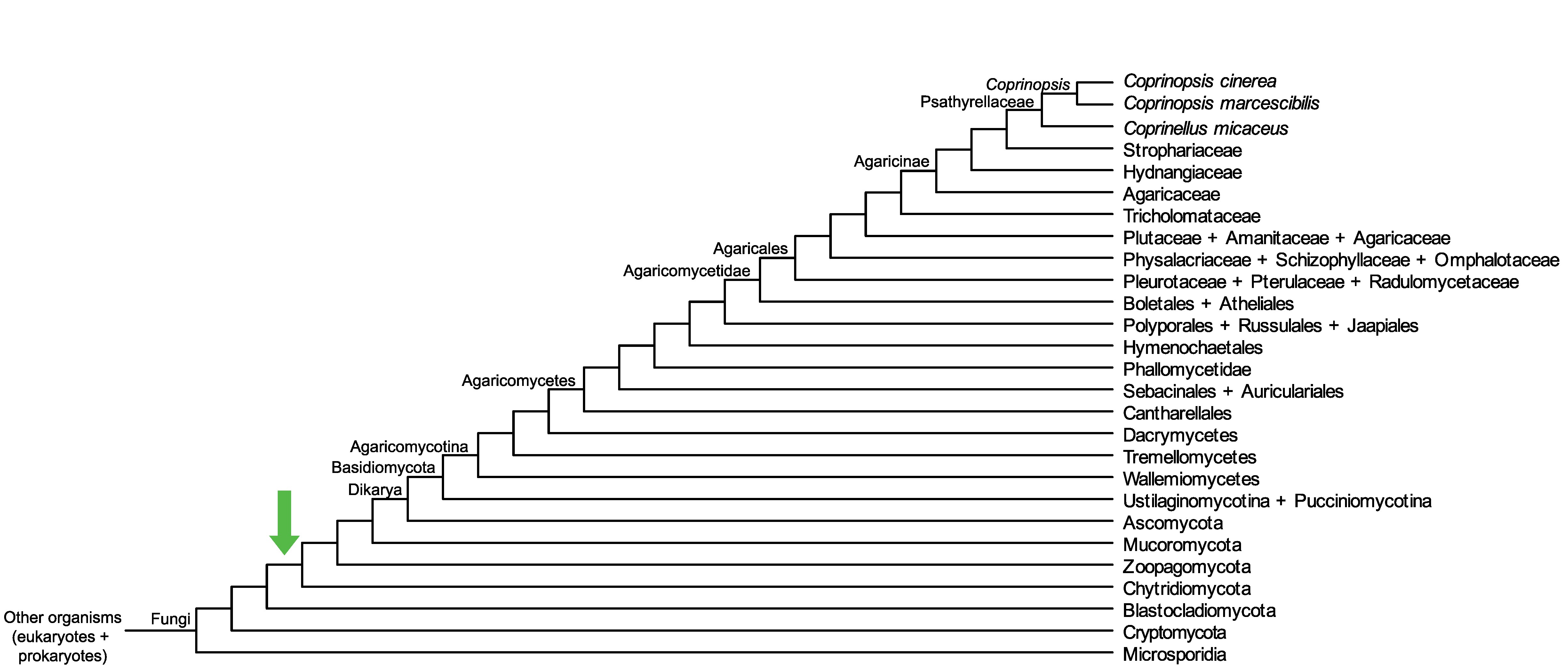
Protein
| Sequence id | CopciAB_376354.T0 |
|---|---|
| Sequence |
>CopciAB_376354.T0 MLILATFSAFLALLPGASALWPIPNDITTGTSPLRLARDFSINLNGVRHAPKDLVDAVSRTQHFLREDKLQLLVP DRGASLKSSISNSPFLKSLTVTLNSRTAKTRSIAEEAIADIGTRQEGYTLTVPADGSEAVLTANSTLGLFRGLTT FSQLWYELDGHVYTVQAPVSIRDAPQYVYRGLMLDTSRNYFPIADIKRTLDAMSWVKVNTLHWHIVDAQSFPLVV PGFEELSRKGAYNPASIYTPNDVKDIVNYAAQRGIDILVEVDTPGHTSIIHHAHPEHIACFEASPWTRYAYEPPA GQLRLASPATVNFTSSLLTSVARLFPSKFFSTGGDEINQPCYEDDAATQKELEKQGKTLEQALDTFTQVTHRALH DMGKTTVVWQEMVLDHKVTLSNDTVAMVWISSQHAKAVAQRGHRLIHAASDYFYLDCGGGGWIGNNPNGNSWCDP FKTWQKAYSFNPRANLTEEEAKLVLGGQQLLWAEQSGPSNLDPIVWPRAAASAEVFWSGHGRDGRTALPRLHDLA YRFVQRGVRAIPLQPQWCALRPGACDIDA |
| Length | 554 |
Coding
| Sequence id | CopciAB_376354.T0 |
|---|---|
| Sequence |
>CopciAB_376354.T0 ATGCTCATCCTCGCCACCTTCTCCGCGTTCTTAGCCTTACTGCCTGGAGCGTCTGCTTTATGGCCTATCCCCAAC GACATCACGACGGGTACGAGCCCGTTGAGGCTCGCTCGAGACTTTTCCATCAACCTGAACGGTGTGCGCCACGCA CCGAAGGACCTGGTCGACGCAGTCTCGCGCACACAGCACTTCCTGAGGGAGGACAAGCTCCAGCTCCTCGTTCCT GATCGCGGTGCATCCCTCAAGAGTTCCATCAGCAACTCGCCTTTCCTCAAGTCGCTCACAGTCACACTGAACAGC AGAACTGCGAAGACCAGGTCCATCGCTGAAGAGGCGATCGCAGATATCGGGACTCGCCAGGAGGGTTATACGCTC ACTGTTCCAGCGGACGGCTCGGAAGCGGTCTTAACTGCCAATTCGACCTTGGGGTTATTCCGTGGTCTGACGACG TTCAGCCAGCTGTGGTATGAGTTGGATGGCCATGTATACACAGTACAGGCTCCTGTCTCTATTCGAGATGCGCCA CAATATGTGTATCGCGGTCTTATGCTCGACACTTCGCGAAACTATTTCCCTATTGCCGACATCAAACGAACGCTG GATGCCATGAGCTGGGTCAAGGTCAACACCCTCCACTGGCACATCGTTGATGCTCAATCCTTCCCTCTCGTTGTT CCTGGGTTCGAGGAATTGTCACGCAAAGGAGCCTACAACCCGGCATCCATCTACACCCCCAATGACGTCAAGGAT ATCGTCAACTACGCTGCTCAGCGAGGAATCGATATTCTCGTTGAAGTAGACACTCCTGGGCACACGTCGATCATC CATCACGCGCATCCCGAACACATCGCTTGTTTCGAGGCCTCCCCCTGGACCCGCTACGCCTACGAACCACCCGCT GGACAGCTACGATTGGCTTCTCCAGCTACTGTCAACTTTACATCCTCCCTTTTGACCTCGGTCGCTAGGTTGTTC CCTTCCAAGTTCTTCAGCACTGGAGGGGATGAGATCAATCAGCCTTGCTATGAAGATGATGCCGCCACCCAGAAG GAATTGGAGAAGCAAGGAAAGACTCTGGAGCAGGCACTCGATACCTTCACTCAAGTTACGCACAGGGCTTTACAT GACATGGGCAAGACAACTGTTGTCTGGCAGGAGATGGTGCTCGACCATAAAGTTACCCTCTCAAACGATACCGTA GCCATGGTATGGATCTCTTCACAGCACGCCAAAGCAGTTGCCCAGCGAGGCCACAGACTCATCCACGCTGCATCC GATTACTTCTACCTCGACTGCGGTGGTGGAGGTTGGATTGGCAACAATCCCAACGGAAACAGCTGGTGTGACCCA TTCAAGACCTGGCAGAAGGCCTATTCTTTCAACCCCAGAGCCAACCTGACTGAAGAGGAAGCGAAGCTGGTTCTT GGAGGCCAACAACTCCTCTGGGCAGAGCAGTCTGGTCCGTCTAACTTGGATCCTATCGTGTGGCCCCGAGCTGCT GCATCCGCAGAGGTATTCTGGTCTGGCCACGGCCGCGATGGTCGAACTGCATTACCGCGCTTGCATGATCTTGCA TACCGTTTCGTTCAACGAGGCGTCCGTGCCATCCCTTTGCAGCCGCAATGGTGTGCCCTTCGCCCTGGTGCATGT GATATCGATGCATGA |
| Length | 1665 |
Transcript
| Sequence id | CopciAB_376354.T0 |
|---|---|
| Sequence |
>CopciAB_376354.T0 ATTCTCCTTACTTACACTCGATGCTCATCCTCGCCACCTTCTCCGCGTTCTTAGCCTTACTGCCTGGAGCGTCTG CTTTATGGCCTATCCCCAACGACATCACGACGGGTACGAGCCCGTTGAGGCTCGCTCGAGACTTTTCCATCAACC TGAACGGTGTGCGCCACGCACCGAAGGACCTGGTCGACGCAGTCTCGCGCACACAGCACTTCCTGAGGGAGGACA AGCTCCAGCTCCTCGTTCCTGATCGCGGTGCATCCCTCAAGAGTTCCATCAGCAACTCGCCTTTCCTCAAGTCGC TCACAGTCACACTGAACAGCAGAACTGCGAAGACCAGGTCCATCGCTGAAGAGGCGATCGCAGATATCGGGACTC GCCAGGAGGGTTATACGCTCACTGTTCCAGCGGACGGCTCGGAAGCGGTCTTAACTGCCAATTCGACCTTGGGGT TATTCCGTGGTCTGACGACGTTCAGCCAGCTGTGGTATGAGTTGGATGGCCATGTATACACAGTACAGGCTCCTG TCTCTATTCGAGATGCGCCACAATATGTGTATCGCGGTCTTATGCTCGACACTTCGCGAAACTATTTCCCTATTG CCGACATCAAACGAACGCTGGATGCCATGAGCTGGGTCAAGGTCAACACCCTCCACTGGCACATCGTTGATGCTC AATCCTTCCCTCTCGTTGTTCCTGGGTTCGAGGAATTGTCACGCAAAGGAGCCTACAACCCGGCATCCATCTACA CCCCCAATGACGTCAAGGATATCGTCAACTACGCTGCTCAGCGAGGAATCGATATTCTCGTTGAAGTAGACACTC CTGGGCACACGTCGATCATCCATCACGCGCATCCCGAACACATCGCTTGTTTCGAGGCCTCCCCCTGGACCCGCT ACGCCTACGAACCACCCGCTGGACAGCTACGATTGGCTTCTCCAGCTACTGTCAACTTTACATCCTCCCTTTTGA CCTCGGTCGCTAGGTTGTTCCCTTCCAAGTTCTTCAGCACTGGAGGGGATGAGATCAATCAGCCTTGCTATGAAG ATGATGCCGCCACCCAGAAGGAATTGGAGAAGCAAGGAAAGACTCTGGAGCAGGCACTCGATACCTTCACTCAAG TTACGCACAGGGCTTTACATGACATGGGCAAGACAACTGTTGTCTGGCAGGAGATGGTGCTCGACCATAAAGTTA CCCTCTCAAACGATACCGTAGCCATGGTATGGATCTCTTCACAGCACGCCAAAGCAGTTGCCCAGCGAGGCCACA GACTCATCCACGCTGCATCCGATTACTTCTACCTCGACTGCGGTGGTGGAGGTTGGATTGGCAACAATCCCAACG GAAACAGCTGGTGTGACCCATTCAAGACCTGGCAGAAGGCCTATTCTTTCAACCCCAGAGCCAACCTGACTGAAG AGGAAGCGAAGCTGGTTCTTGGAGGCCAACAACTCCTCTGGGCAGAGCAGTCTGGTCCGTCTAACTTGGATCCTA TCGTGTGGCCCCGAGCTGCTGCATCCGCAGAGGTATTCTGGTCTGGCCACGGCCGCGATGGTCGAACTGCATTAC CGCGCTTGCATGATCTTGCATACCGTTTCGTTCAACGAGGCGTCCGTGCCATCCCTTTGCAGCCGCAATGGTGTG CCCTTCGCCCTGGTGCATGTGATATCGATGCATGATATATGTTATGCATTGACGAGCGCTCCTCGAAGGACAATT ATACGGTCTGGATGAATTTGGAAACACGAACGCTCGATTGCAGGATATGTTTGGACATCATACCTTGAATGTAAT AGCTATTTTCAAATTAGCTCGAATTGCTTG |
| Length | 1830 |
Gene
| Sequence id | CopciAB_376354.T0 |
|---|---|
| Sequence |
>CopciAB_376354.T0 ATTCTCCTTACTTACACTCGATGCTCATCCTCGCCACCTTCTCCGCGTTCTTAGCCTTACTGCCTGGAGCGTCTG CTTTATGGCCTATCCCCAACGACATCACGACGGGTACGAGCCCGTTGAGGCTCGCTCGAGACTTTTCCATCAACC TGAACGGTGTGCGCCACGCACCGAAGGACCTGGTCGACGCAGTCTCGCGCACACAGCACTTCCTGAGGGAGGACA AGCTCCAGCTCCTCGTTCCTGATCGCGGTGCATCCCTCAAGAGTTCCATCAGCAACTCGCCTTTCCTCAAGTCGC TCACAGTCACACTGAACAGCAGAACTGCGAAGACCAGGTCCATCGCTGAAGAGGCGATCGCAGATATCGGGACTC GCCAGGAGGGTTATACGCTCACTGTTCCAGCGGACGGCTCGGAAGCGGTCTTAACTGCCAATTCGACCTTGGGGT TATTCCGTGGTCTGACGACGTTCAGCCAGCTGTGGTATGAGTTGGATGGCCATGTATACACAGTACAGGCTCCTG TCTCTATTCGAGATGCGCCACAATATGTGCGTGCAGTTTCTTTTCATGAATTCAGTGTGGATACTCACAGGCCGT TTTGACATGCAGGTGTATCGCGGTCTTATGCTCGACACTTCGCGAAACTAGTGAGAGGCTTCCCCAGTCCCTCAG TACAGGATGCTCACGTCTCTCAGTTTCCCTATTGCCGACATCAAACGAACGCTGGATGCCATGAGCTGGGTCAAG GTGTGTTTAAACAACGTTTTGACGAGCTTGATACAAATTCATGGCAGGTCAACACCCTCCACTGGCACATCGTTG ATGCTCAATCCTTCCCTCTCGTTGTTCCTGGGTTCGAGGAATTGTCACGCAAAGGAGCCTACAACCCGGCATCCA TCTACACCCCCAATGACGTCAAGGATATCGTCAACTACGCTGCTCAGCGAGGAATCGATATTCTCGTTGAAGTAG ACACTCCTGGGCACACGTCGATCATCCATCACGCGCATCCCGAACACATCGCTTGTTTCGAGGCCTCCCCCTGGA CCCGCTACGCCTACGGCAAGTGTATTCTTTTGTGCGGCGTGTCTTTCTCATCCCAAACCCCGTCCACTAGAACCA CCCGCTGGACAGCTACGATTGGCTTCTCCAGCTACTGTCAACTTTACATCCTCCCTTTTGACCTCGGTCGCTAGG TTGTTCCCTTCCAAGTTCTTCAGCACTGGAGGGGATGAGATCAATCAGCCTTGCTATGAAGATGATGCCGCCACC CAGAAGGAATTGGGTATGTTGCATTTTATGTGAATGGATAAGACCCTGATTATCTCGGACTTGTAGAGAAGCAAG GAAAGACTCTGGAGCAGGCACTCGATACCTTCACTCAAGTTACGCACAGGGCTTTACATGACATGGGCAAGACAA CTGTTGTCTGGCAGGGTATGTCTCATACTTACTCCTACGACCGCGTTGAGAACTCAAGCAGCGCTGTAGAGATGG TGCTCGACCATAAAGTTACCCTCTCAAACGATACCGTAGCCATGTATGTCCTTCCACTGACCTACACCACACCCA CCGAACGTTCATATTGCTCTTTCCAGGGTATGGATCTCTTCACAGCACGCCAAAGCAGTTGCCCAGCGAGGCCAC AGACTCATCCACGCTGCATCCGATTACTTCTACCTCGACTGCGGTGGTGGAGGTTGGATTGGCAACAATCCCAAC GGAAACAGCTGGTGTGACCCATTCAAGACCTGGCAGAAGGCCTATTCTTTCAACCCCAGAGCCAACCTGACTGAA GAGGAAGCGAAGCTGGTTCTTGGAGGCCAACAACTCCTCTGGGCAGAGCAGTCTGGTCCGTCTAACTTGGATCCT ATCGTGTGGCCCCGAGCTGCTGCATCCGCAGAGGTGAGTTTAGATAGATACAAACGACGTGGGCTTGGAACTGAA ACCTGATGCGCTGTACTCTAGGTATTCTGGTCTGGCCACGGCCGCGATGGTCGAACTGCATTACCGCGCTTGCAT GATCTTGCATACCGTTTCGTTCAACGAGGCGTCCGTGCCATCCCTTTGCAGCCGCAATGGTGTGCCCTTCGCCCT GGTGCATGTGATATCGATGCATGATATATGTTATGCATTGACGAGCGCTCCTCGAAGGACAATTATACGGTCTGG ATGAATTTGGAAACACGAACGCTCGATTGCAGGATATGTTTGGACATCATACCTTGAATGTAATAGCTATTTTCA AATTAGCTCGAATTGCTTG |
| Length | 2269 |