CopciAB_377156
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_377156 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 377156 |
| Uniprot id | Functional description | Belongs to the glycosyl hydrolase 5 (cellulase A) family | |
| Location | scaffold_2:679414..681323 | Strand | + |
| Gene length (nt) | 1910 | Transcript length (nt) | 1632 |
| CDS length (nt) | 1446 | Protein length (aa) | 481 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agrocybe aegerita | Agrae_CAA7270937 | 74 | 2.344E-258 | 792 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_92417 | 69.6 | 1.342E-245 | 755 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB21881 | 71.9 | 3.817E-245 | 754 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1079888 | 68 | 1.153E-237 | 732 |
| Grifola frondosa | Grifr_OBZ72767 | 66.5 | 6.077E-237 | 730 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1447403 | 67.8 | 6.679E-237 | 730 |
| Pleurotus ostreatus PC9 | PleosPC9_1_64780 | 66.8 | 6.643E-235 | 724 |
| Lentinula edodes B17 | Lened_B_1_1_12316 | 66.5 | 1.52E-233 | 720 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_16912 | 65.3 | 2.084E-232 | 717 |
| Lentinula edodes NBRC 111202 | Lenedo1_1059714 | 65.4 | 5.093E-231 | 713 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_77559 | 64.8 | 1.243E-226 | 700 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_188761 | 65 | 2.646E-226 | 699 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_377156.T0 |
| Description | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00150 | Cellulase (glycosyl hydrolase family 5) | IPR001547 | 81 | 343 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR001547 | Glycoside hydrolase, family 5 |
| IPR017853 | Glycoside hydrolase superfamily |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds | MF |
| GO:0071704 | organic substance metabolic process | BP |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| G | Belongs to the glycosyl hydrolase 5 (cellulase A) family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| GH | GH5 | GH5_22 |
Transcription factor
| Group |
|---|
| No records |
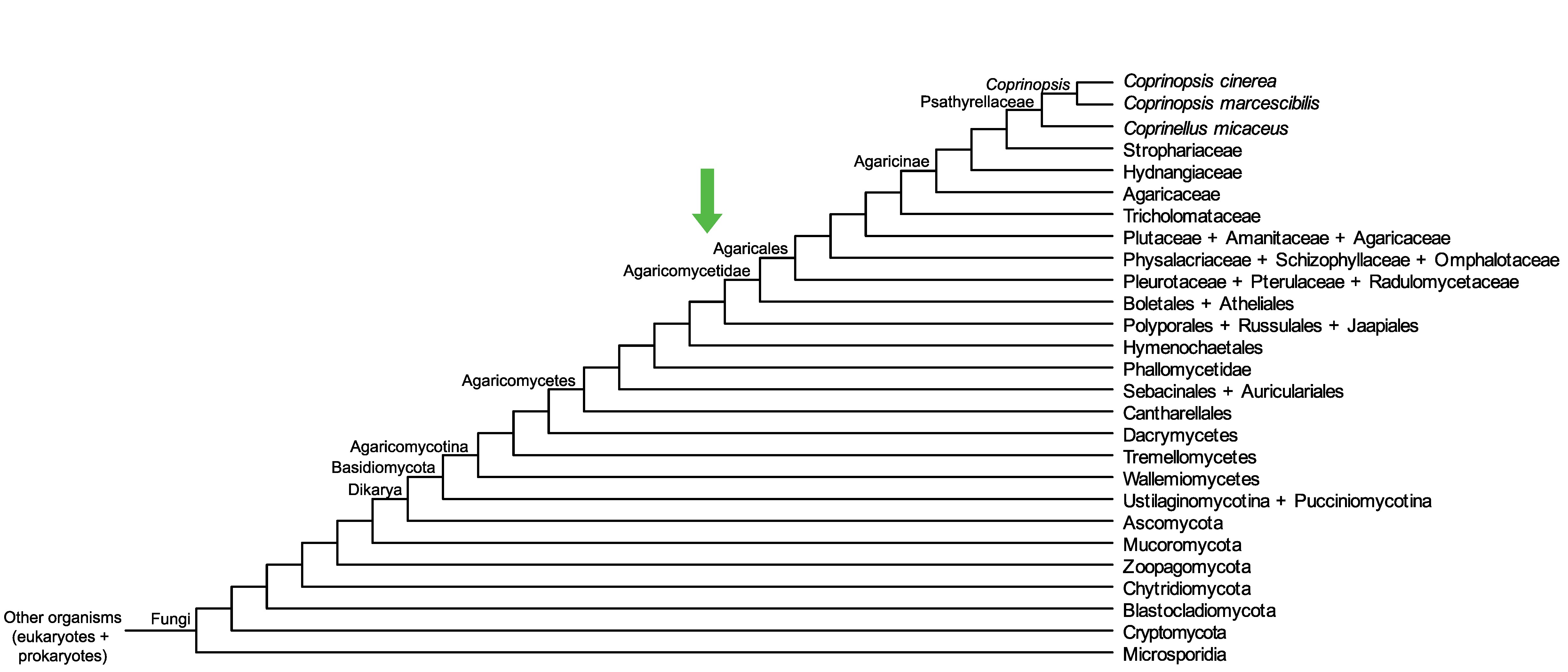
Conservation of CopciAB_377156 across fungi.
Arrow shows the origin of gene family containing CopciAB_377156.
Protein
| Sequence id | CopciAB_377156.T0 |
|---|---|
| Sequence |
>CopciAB_377156.T0 MAAGAFLKVDGTRIVDQDNNEVVLHGAGLGGWMTMENFISGFPACEFQVRDALEETIGKEKAAFFFDKFLEHFFT EKDAIFFKSLGLNCIRIAVGYRHFEDDMNPRVLKPDAFKHLDRAISLCAKHSIYTVIDVHTAPGGQSGGWHADAG VHIANFWRHKDFQDRLVWLWTELAKHYKDNPWIAGYNVLNEPADPHPQHAGLIKMYDRLHQAIREIDGNHIIFLD GNTFATDFTKFPEDAGTRWTNTAYAIHDYAVYGFPSAPEPYEGSEAQKERLLKTYKRKREWMDQRGLCVWNGEWG PVYARREYDGDAMEDINERRYNVLKDQLEIYEKDRLSWSIWLYKDIGFQGMVYVSPDTPYRQRFKDFLAKKHRLA ADAWGKDDQHVKQFYSPIISLIEDNIKDKSHLKLYPPLWTVPERTTRLARTMLVAEYLVQEWADLFLGLDEQQLE ELAKSFSFDNCLKRDGLNEVLTAHAERVSKK |
| Length | 481 |
Coding
| Sequence id | CopciAB_377156.T0 |
|---|---|
| Sequence |
>CopciAB_377156.T0 ATGGCTGCAGGCGCGTTCTTGAAAGTGGATGGAACCCGGATTGTGGACCAGGACAACAATGAGGTTGTGTTGCAT GGGGCTGGGCTTGGTGGTTGGATGACGATGGAGAACTTCATTTCAGGTTTCCCGGCCTGTGAGTTCCAGGTACGC GATGCGTTGGAGGAGACCATCGGCAAGGAGAAGGCCGCTTTCTTCTTTGACAAGTTCCTGGAGCATTTCTTCACG GAGAAGGATGCCATCTTCTTCAAGTCTTTGGGCTTGAACTGCATTCGAATCGCAGTGGGGTACCGACACTTTGAA GACGACATGAACCCCCGCGTCCTCAAACCAGACGCCTTCAAGCACCTCGACAGGGCCATCTCCCTCTGCGCCAAA CACTCCATCTACACCGTCATCGACGTGCACACCGCACCCGGCGGGCAGAGCGGGGGCTGGCACGCCGACGCCGGC GTGCACATCGCCAACTTCTGGAGACACAAAGACTTCCAAGACCGGCTGGTCTGGCTCTGGACCGAGCTCGCAAAG CACTACAAAGACAACCCCTGGATCGCGGGCTACAACGTCCTCAACGAGCCCGCGGACCCGCACCCCCAGCACGCG GGTTTGATCAAGATGTACGACAGGCTGCATCAGGCTATTCGCGAGATTGATGGGAACCATATCATCTTCCTCGAT GGGAACACGTTTGCGACGGATTTTACAAAGTTTCCCGAGGATGCGGGGACGAGGTGGACGAATACGGCGTATGCG ATACATGATTACGCTGTTTATGGGTTTCCGAGTGCGCCGGAGCCGTATGAGGGCTCCGAGGCCCAGAAGGAGAGG TTGCTAAAGACGTATAAGAGGAAGAGGGAGTGGATGGATCAGAGGGGTTTGTGTGTGTGGAATGGGGAGTGGGGG CCTGTGTATGCGAGGAGGGAGTATGATGGCGATGCGATGGAGGATATTAATGAGAGGAGATATAACGTGTTGAAG GACCAATTGGAGATTTATGAAAAGGATCGCCTCAGCTGGTCCATCTGGCTCTATAAAGACATTGGCTTCCAAGGG ATGGTCTACGTCTCCCCAGACACGCCCTACCGCCAACGCTTCAAAGACTTCCTCGCCAAGAAACACAGACTCGCA GCAGACGCCTGGGGTAAAGACGACCAACACGTCAAGCAATTCTATAGCCCCATCATCTCCTTGATCGAGGACAAC ATCAAGGATAAATCCCACCTCAAGCTCTACCCGCCATTGTGGACCGTTCCGGAGAGGACGACAAGACTGGCGAGA ACGATGTTGGTGGCAGAATACCTGGTGCAAGAGTGGGCAGACTTGTTCCTGGGATTGGACGAACAACAGCTGGAG GAACTAGCGAAGAGTTTTTCGTTCGATAACTGCTTGAAGCGGGATGGGTTGAACGAGGTTCTAACGGCTCATGCC GAAAGGGTCAGCAAGAAATAG |
| Length | 1446 |
Transcript
| Sequence id | CopciAB_377156.T0 |
|---|---|
| Sequence |
>CopciAB_377156.T0 AAAGGTGACCCTGCTCGATCACACTTAATTACGACCATGGCTGCAGGCGCGTTCTTGAAAGTGGATGGAACCCGG ATTGTGGACCAGGACAACAATGAGGTTGTGTTGCATGGGGCTGGGCTTGGTGGTTGGATGACGATGGAGAACTTC ATTTCAGGTTTCCCGGCCTGTGAGTTCCAGGTACGCGATGCGTTGGAGGAGACCATCGGCAAGGAGAAGGCCGCT TTCTTCTTTGACAAGTTCCTGGAGCATTTCTTCACGGAGAAGGATGCCATCTTCTTCAAGTCTTTGGGCTTGAAC TGCATTCGAATCGCAGTGGGGTACCGACACTTTGAAGACGACATGAACCCCCGCGTCCTCAAACCAGACGCCTTC AAGCACCTCGACAGGGCCATCTCCCTCTGCGCCAAACACTCCATCTACACCGTCATCGACGTGCACACCGCACCC GGCGGGCAGAGCGGGGGCTGGCACGCCGACGCCGGCGTGCACATCGCCAACTTCTGGAGACACAAAGACTTCCAA GACCGGCTGGTCTGGCTCTGGACCGAGCTCGCAAAGCACTACAAAGACAACCCCTGGATCGCGGGCTACAACGTC CTCAACGAGCCCGCGGACCCGCACCCCCAGCACGCGGGTTTGATCAAGATGTACGACAGGCTGCATCAGGCTATT CGCGAGATTGATGGGAACCATATCATCTTCCTCGATGGGAACACGTTTGCGACGGATTTTACAAAGTTTCCCGAG GATGCGGGGACGAGGTGGACGAATACGGCGTATGCGATACATGATTACGCTGTTTATGGGTTTCCGAGTGCGCCG GAGCCGTATGAGGGCTCCGAGGCCCAGAAGGAGAGGTTGCTAAAGACGTATAAGAGGAAGAGGGAGTGGATGGAT CAGAGGGGTTTGTGTGTGTGGAATGGGGAGTGGGGGCCTGTGTATGCGAGGAGGGAGTATGATGGCGATGCGATG GAGGATATTAATGAGAGGAGATATAACGTGTTGAAGGACCAATTGGAGATTTATGAAAAGGATCGCCTCAGCTGG TCCATCTGGCTCTATAAAGACATTGGCTTCCAAGGGATGGTCTACGTCTCCCCAGACACGCCCTACCGCCAACGC TTCAAAGACTTCCTCGCCAAGAAACACAGACTCGCAGCAGACGCCTGGGGTAAAGACGACCAACACGTCAAGCAA TTCTATAGCCCCATCATCTCCTTGATCGAGGACAACATCAAGGATAAATCCCACCTCAAGCTCTACCCGCCATTG TGGACCGTTCCGGAGAGGACGACAAGACTGGCGAGAACGATGTTGGTGGCAGAATACCTGGTGCAAGAGTGGGCA GACTTGTTCCTGGGATTGGACGAACAACAGCTGGAGGAACTAGCGAAGAGTTTTTCGTTCGATAACTGCTTGAAG CGGGATGGGTTGAACGAGGTTCTAACGGCTCATGCCGAAAGGGTCAGCAAGAAATAGTAGATGGGTATGGAATGC AGTCTCTAAAATACATGACAACAGTGTAGGAGAGGATGTTACAACAACAGTACTGTACAACTAGCGTAGGGGATA GCACGCGAAGCAAGAAATATGGACAACAAAGAATGAACAACGAACGATAATCCGAAA |
| Length | 1632 |
Gene
| Sequence id | CopciAB_377156.T0 |
|---|---|
| Sequence |
>CopciAB_377156.T0 AAAGGTGACCCTGCTCGATCACACTTAATTACGACCATGGCTGCAGGCGCGTTCTTGAAAGTGGATGGAACCCGG ATTGTGGACCAGGACAACAATGAGGTTGTGTTGCATGGGGCTGGGCTTGGTGGTTGGATGACGTGAGTCACCGTT TGATATGCTGCGGGTGCGAAGGCTGAATGAGCCGGTTAGGATGGAGAACTTCATTTCAGGTATTGCAAGGCAGAC TTGACGCTGCGGGAAAGTTAATCTTTGGGCCTCGTTGACTAGGTTTCCCGGCCTGTGAGTTCCAGGTACGCGATG CGTTGGAGGAGACCATCGGCAAGGAGAAGGCCGCTTTCTTCTTTGACAAGGTACACTCTGACGAAGGAATGACCG TCAGTCTAGCTGACAAGGCAGTGTCGTAGTTCCTGGAGCATTTCTTCACGGAGAAGGATGCCATCTTCTTCAAGT CTTTGGGCTTGAACTGCATTCGAATCGCAGTGGGGTACCGACACTTTGAAGGTGCGCATCACTAGCCCCTATCCC CGCATTTAAATGACCACTCACCGTGCTACACCCAGACGACATGAACCCCCGCGTCCTCAAACCAGACGCCTTCAA GCACCTCGACAGGGCCATCTCCCTCTGCGCCAAACACTCCATCTACACCGTCATCGACGTGCACACCGCACCCGG CGGGCAGAGCGGGGGCTGGCACGCCGACGCCGGCGTGCACATCGCCAACTTCTGGAGACACAAAGACTTCCAAGA CCGGCTGGTCTGGCTCTGGACCGAGCTCGCAAAGCACTACAAAGACAACCCCTGGATCGCGGGCTACAACGTCCT CAACGAGCCCGCGGACCCGCACCCCCAGCACGCGGGTTTGATCAAGATGTACGACAGGCTGCATCAGGCTATTCG CGAGATTGATGGGAACCATATCATCTTCCTCGATGGGAACACGTTTGCGACGGATTTTACAAAGTTTCCCGAGGA TGCGGGGACGAGGTGGACGAATACGGCGTATGCGATACATGATTACGCTGTTTATGGGTTTCCGAGTGCGCCGGA GCCGTATGAGGGCTCCGAGGCCCAGAAGGAGAGGTTGCTAAAGACGTATAAGAGGAAGAGGGAGTGGATGGATCA GAGGGGTTTGTGTGTGTGGAATGGGGAGTGGGGGCCTGTGTATGCGAGGAGGGAGTATGATGGCGATGCGATGGA GGATATTAATGAGAGGAGATATAACGTGTTGAAGGACCAATTGGAGATTTATGAAAAGGTAATTTACATGTACTT TGCGTTCGTTGTGCGAGGCTGACGGGTGTCGACTCTAGGATCGCCTCAGCTGGTCCATCTGGCTCTATAAAGACA TTGGCTTCCAAGGGATGGTCTACGTCTCCCCAGACACGCCCTACCGCCAACGCTTCAAAGACTTCCTCGCCAAGA AACACAGACTCGCAGCAGACGCCTGGGGTAAAGACGACCAACACGTCAAGCAATTCTATAGCCCCATCATCTCCT TGATCGAGGACAACATCAAGGATAAATCCCACCTCAAGCTCTACCCGCCATTGTGGACCGTTCCGGAGAGGACGA CAAGACTGGCGAGAACGATGTTGGTGGCAGAATACCTGGTGCAAGAGTGGGCAGACTTGTTCCTGGGATTGGACG AACAACAGCTGGAGGAACTAGCGAAGAGTTTTTCGTTCGATAACTGCTTGAAGCGGGATGGGTTGAACGAGGTTC TAACGGCTCATGCCGAAAGGGTCAGCAAGAAATAGTAGATGGGTATGGAATGCAGTCTCTAAAATACATGACAAC AGTGTAGGAGAGGATGTTACAACAACAGTACTGTACAACTAGCGTAGGGGATAGCACGCGAAGCAAGAAATATGG ACAACAAAGAATGAACAACGAACGATAATCCGAAA |
| Length | 1910 |