CopciAB_381351
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_381351 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 381351 |
| Uniprot id | Functional description | Belongs to the GMC oxidoreductase family | |
| Location | scaffold_6:2593760..2596588 | Strand | - |
| Gene length (nt) | 2829 | Transcript length (nt) | 2513 |
| CDS length (nt) | 2088 | Protein length (aa) | 695 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN4006 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_105031 | 48.8 | 1.461E-208 | 659 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB27685 | 46.8 | 2.602E-206 | 653 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205329 | 46.7 | 7.998E-206 | 651 |
| Auricularia subglabra | Aurde3_1_1236104 | 45.6 | 4.133E-183 | 586 |
| Schizophyllum commune H4-8 | Schco3_2511180 | 45.9 | 6.875E-182 | 582 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1451848 | 45.2 | 7.056E-180 | 576 |
| Pleurotus ostreatus PC9 | PleosPC9_1_117616 | 45.1 | 5.872E-180 | 576 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1088797 | 45 | 1.423E-178 | 572 |
| Flammulina velutipes | Flave_chr08AA01120 | 45.3 | 1.224E-170 | 549 |
| Lentinula edodes NBRC 111202 | Lenedo1_1061718 | 43 | 1.326E-170 | 549 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_4405 | 43 | 2.902E-170 | 548 |
| Lentinula edodes B17 | Lened_B_1_1_3883 | 42.8 | 9.332E-149 | 485 |
| Grifola frondosa | Grifr_OBZ73613 | 35.7 | 3.393E-112 | 378 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_381351.T0 |
| Description | Belongs to the GMC oxidoreductase family |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF00732 | GMC oxidoreductase | IPR000172 | 49 | 372 |
| Pfam | PF05199 | GMC oxidoreductase | IPR007867 | 495 | 632 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| SP(Sec/SPI) | 1 | 20 | 0.9644 |
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR000172 | Glucose-methanol-choline oxidoreductase, N-terminal |
| IPR007867 | Glucose-methanol-choline oxidoreductase, C-terminal |
| IPR027424 | Glucose Oxidase, domain 2 |
| IPR036188 | FAD/NAD(P)-binding domain superfamily |
| IPR012132 | Glucose-methanol-choline oxidoreductase |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors | MF |
| GO:0050660 | flavin adenine dinucleotide binding | MF |
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| E | Belongs to the GMC oxidoreductase family |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
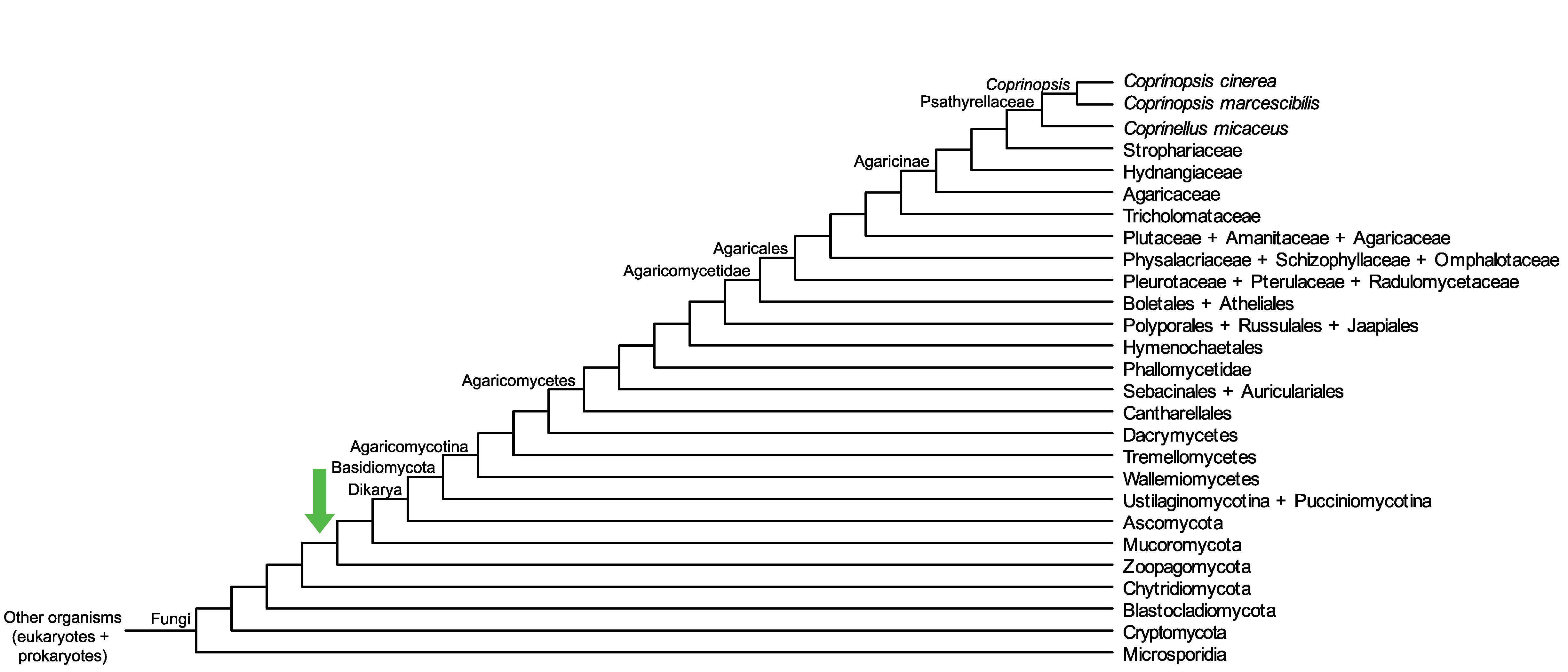
Conservation of CopciAB_381351 across fungi.
Arrow shows the origin of gene family containing CopciAB_381351.
Protein
| Sequence id | CopciAB_381351.T0 |
|---|---|
| Sequence |
>CopciAB_381351.T0 MLSLELLVLTLTALTAPVLAHRDYNNVHARRFGLTPRDYITPETRLESYDYIIAGGGLAGLVLASKLSADGETTV LVLEAGGTGDEQRETIDRPGLTYFRSLLNTEHDYAYKTTPQDGAGGRIMNWPRGKILGGSSAVNGMYLVRPNALE VNAMHEMNADNDAKEYADAWTWDSLLEAMKQGETFTPPTQEALDVAGMRFNPDSHGSSGPLQATYSAYMVPISSA WLPTLEAAGVPISNDAYSGDNVGGFFSIMAINPSNWTRSYAKSAYIDNLPPRSNLHILTNSAVTRILFADNAQDG LQVASGVEFASDANDTPKTVLANKEVILAGGAVGSPHMLLVSGVGDRALLESLDIPVRVDLPGVGHHMQDHLAVS VAWSTEEDTQGTIFESGSEFSQSPEFLSFVNSGTSYVNGRFLFDGQEQFDAYLQGLRDTFSSADDLAPLLPGTSD EVDEGYRTIYQTLVDKVYPEAGLVEMLYSINAAGRIIVQAAIQTPLSQGRLSINSTSIFDPPVVDPKYFSHPGDI VLMRQGVKQVRRMSTLSPLRDILGDELSPGPDVQSDEDIENWIRQNANTEFHPGCTCVMLPRDKGGVIDQSLKVH GTANLRVIDGSIFPLSMSAHLMAPIYGVAEKAAELILNPPSSSGGGSGGKGGSSGGKGGSGKNDEDDDESAAAGL SSSLFITVAGSLFTALLGAF |
| Length | 695 |
Coding
| Sequence id | CopciAB_381351.T0 |
|---|---|
| Sequence |
>CopciAB_381351.T0 ATGCTGTCTCTTGAACTTCTCGTTCTTACGCTAACGGCACTAACCGCCCCTGTTCTCGCCCATCGCGACTACAAC AATGTTCACGCCCGACGGTTTGGTCTAACCCCTCGGGACTACATCACCCCCGAAACACGACTCGAATCCTACGAC TATATCATTGCCGGAGGAGGTCTTGCAGGACTGGTACTTGCTTCCAAGCTCTCGGCTGATGGGGAAACCACTGTT CTCGTCCTCGAGGCTGGAGGAACAGGCGACGAGCAGAGAGAGACTATTGACCGTCCTGGCTTGACATACTTCCGC TCTCTCCTCAACACCGAGCACGATTATGCCTACAAGACCACCCCTCAGGACGGTGCTGGTGGTCGTATCATGAAC TGGCCTCGTGGAAAGATCCTTGGAGGCTCTTCTGCGGTTAACGGCATGTATCTCGTTCGACCCAACGCGCTCGAA GTTAATGCCATGCACGAGATGAACGCCGATAACGACGCCAAGGAATATGCTGACGCGTGGACCTGGGACTCCCTT CTCGAAGCTATGAAGCAGGGCGAGACCTTCACCCCTCCCACCCAGGAAGCCCTCGATGTTGCGGGCATGCGCTTC AACCCGGACAGCCATGGCTCCAGCGGACCTCTGCAGGCCACTTACTCTGCCTACATGGTCCCCATCTCTAGTGCC TGGCTCCCGACCCTCGAGGCTGCTGGTGTTCCCATTTCCAATGACGCCTACTCCGGAGACAACGTTGGCGGCTTC TTCAGTATCATGGCGATCAACCCCTCCAACTGGACTCGATCATACGCCAAGTCTGCCTATATCGACAACCTTCCT CCCCGCTCCAACCTCCACATCCTCACCAACAGTGCCGTCACCCGTATCCTCTTTGCCGACAATGCTCAAGATGGC CTCCAGGTCGCGAGCGGAGTCGAGTTCGCCAGCGATGCCAATGATACTCCCAAGACTGTTCTCGCCAACAAGGAA GTGATTCTCGCTGGTGGTGCCGTCGGCAGCCCACACATGTTGTTGGTCAGCGGTGTTGGTGACAGGGCACTGCTT GAGAGCTTGGACATCCCCGTTCGTGTGGACCTCCCTGGTGTTGGCCACCACATGCAAGACCATTTGGCCGTCAGC GTCGCCTGGAGCACTGAGGAAGACACCCAAGGCACCATCTTCGAATCCGGATCCGAGTTCTCTCAATCCCCCGAG TTCCTTTCCTTCGTAAACTCCGGTACTTCCTATGTCAACGGACGATTCCTGTTCGACGGCCAGGAGCAATTCGAC GCCTACCTCCAAGGTCTCCGTGACACCTTCAGCAGCGCCGATGATCTCGCTCCCCTCCTCCCAGGCACCAGCGAC GAGGTTGATGAGGGTTACAGGACCATCTACCAGACTTTGGTCGACAAGGTCTACCCCGAGGCTGGCCTGGTCGAG ATGCTTTACAGCATCAACGCTGCCGGTCGCATCATCGTCCAGGCTGCAATCCAGACCCCTCTGAGCCAAGGCCGC CTCTCCATCAACTCGACCTCGATCTTCGACCCGCCCGTCGTTGACCCCAAGTACTTCTCCCACCCCGGCGATATC GTCCTCATGCGCCAAGGTGTGAAGCAGGTCCGCCGCATGTCGACCCTCTCACCCCTCCGTGATATCCTTGGCGAC GAGCTCAGCCCCGGTCCTGATGTCCAGTCCGACGAGGACATCGAGAACTGGATCAGGCAGAATGCCAACACCGAG TTCCACCCCGGCTGCACTTGCGTGATGTTGCCCCGCGACAAGGGCGGTGTTATCGACCAGTCCCTCAAGGTTCAC GGTACCGCCAACCTCCGCGTTATTGATGGATCCATCTTCCCTCTCTCTATGTCCGCTCACTTGATGGCCCCGATC TACGGTGTCGCCGAGAAGGCTGCCGAACTCATCCTCAACCCTCCCTCGTCATCCGGCGGTGGCTCTGGTGGCAAG GGCGGCTCCTCTGGTGGCAAAGGTGGATCCGGCAAGAACGACGAAGATGACGACGAGAGCGCAGCTGCTGGTCTT TCCAGCTCGCTCTTCATCACTGTCGCTGGTTCCCTCTTCACCGCTCTCCTCGGCGCTTTCTAA |
| Length | 2088 |
Transcript
| Sequence id | CopciAB_381351.T0 |
|---|---|
| Sequence |
>CopciAB_381351.T0 ACCACAACAAACACTTCAGTTTTCGTCCAAACGCACCTTTATCCACTGGCGGGAGACGTTCTGAACCTCCCTCCC TTTCGTTCTTTACCCTAAATCACCTTTTTCTTCCTCCTTTCCGCCCGATGCTGTCTCTTGAACTTCTCGTTCTTA CGCTAACGGCACTAACCGCCCCTGTTCTCGCCCATCGCGACTACAACAATGTTCACGCCCGACGGTTTGGTCTAA CCCCTCGGGACTACATCACCCCCGAAACACGACTCGAATCCTACGACTATATCATTGCCGGAGGAGGTCTTGCAG GACTGGTACTTGCTTCCAAGCTCTCGGCTGATGGGGAAACCACTGTTCTCGTCCTCGAGGCTGGAGGAACAGGCG ACGAGCAGAGAGAGACTATTGACCGTCCTGGCTTGACATACTTCCGCTCTCTCCTCAACACCGAGCACGATTATG CCTACAAGACCACCCCTCAGGACGGTGCTGGTGGTCGTATCATGAACTGGCCTCGTGGAAAGATCCTTGGAGGCT CTTCTGCGGTTAACGGCATGTATCTCGTTCGACCCAACGCGCTCGAAGTTAATGCCATGCACGAGATGAACGCCG ATAACGACGCCAAGGAATATGCTGACGCGTGGACCTGGGACTCCCTTCTCGAAGCTATGAAGCAGGGCGAGACCT TCACCCCTCCCACCCAGGAAGCCCTCGATGTTGCGGGCATGCGCTTCAACCCGGACAGCCATGGCTCCAGCGGAC CTCTGCAGGCCACTTACTCTGCCTACATGGTCCCCATCTCTAGTGCCTGGCTCCCGACCCTCGAGGCTGCTGGTG TTCCCATTTCCAATGACGCCTACTCCGGAGACAACGTTGGCGGCTTCTTCAGTATCATGGCGATCAACCCCTCCA ACTGGACTCGATCATACGCCAAGTCTGCCTATATCGACAACCTTCCTCCCCGCTCCAACCTCCACATCCTCACCA ACAGTGCCGTCACCCGTATCCTCTTTGCCGACAATGCTCAAGATGGCCTCCAGGTCGCGAGCGGAGTCGAGTTCG CCAGCGATGCCAATGATACTCCCAAGACTGTTCTCGCCAACAAGGAAGTGATTCTCGCTGGTGGTGCCGTCGGCA GCCCACACATGTTGTTGGTCAGCGGTGTTGGTGACAGGGCACTGCTTGAGAGCTTGGACATCCCCGTTCGTGTGG ACCTCCCTGGTGTTGGCCACCACATGCAAGACCATTTGGCCGTCAGCGTCGCCTGGAGCACTGAGGAAGACACCC AAGGCACCATCTTCGAATCCGGATCCGAGTTCTCTCAATCCCCCGAGTTCCTTTCCTTCGTAAACTCCGGTACTT CCTATGTCAACGGACGATTCCTGTTCGACGGCCAGGAGCAATTCGACGCCTACCTCCAAGGTCTCCGTGACACCT TCAGCAGCGCCGATGATCTCGCTCCCCTCCTCCCAGGCACCAGCGACGAGGTTGATGAGGGTTACAGGACCATCT ACCAGACTTTGGTCGACAAGGTCTACCCCGAGGCTGGCCTGGTCGAGATGCTTTACAGCATCAACGCTGCCGGTC GCATCATCGTCCAGGCTGCAATCCAGACCCCTCTGAGCCAAGGCCGCCTCTCCATCAACTCGACCTCGATCTTCG ACCCGCCCGTCGTTGACCCCAAGTACTTCTCCCACCCCGGCGATATCGTCCTCATGCGCCAAGGTGTGAAGCAGG TCCGCCGCATGTCGACCCTCTCACCCCTCCGTGATATCCTTGGCGACGAGCTCAGCCCCGGTCCTGATGTCCAGT CCGACGAGGACATCGAGAACTGGATCAGGCAGAATGCCAACACCGAGTTCCACCCCGGCTGCACTTGCGTGATGT TGCCCCGCGACAAGGGCGGTGTTATCGACCAGTCCCTCAAGGTTCACGGTACCGCCAACCTCCGCGTTATTGATG GATCCATCTTCCCTCTCTCTATGTCCGCTCACTTGATGGCCCCGATCTACGGTGTCGCCGAGAAGGCTGCCGAAC TCATCCTCAACCCTCCCTCGTCATCCGGCGGTGGCTCTGGTGGCAAGGGCGGCTCCTCTGGTGGCAAAGGTGGAT CCGGCAAGAACGACGAAGATGACGACGAGAGCGCAGCTGCTGGTCTTTCCAGCTCGCTCTTCATCACTGTCGCTG GTTCCCTCTTCACCGCTCTCCTCGGCGCTTTCTAAAGCTCCTCCTGTCTGACTGACTGACTGACTAACTAACGAT ATGGACGTTCTATGACCCTTCGGATTCCCATTGCTGGATTTGTTTCTTGACCACGACACTCATTTGGCTGTCTTA TGGACTTGACTTGATACTCGTATATCTTTTCGTTCATTCCTAGCCTAATGAACAGGGACTTTCCATGCAACCTTT TGATACCATCGCTGTAGATGACGCTGTTATATTAGAGAAAGATGTAATACTGTTGCAAAGTAGTTACCCCCCTGT GTAGCATAGGAATGAACGGACTAGTGTTTGATTTTTGG |
| Length | 2513 |
Gene
| Sequence id | CopciAB_381351.T0 |
|---|---|
| Sequence |
>CopciAB_381351.T0 ACCACAACAAACACTTCAGTTTTCGTCCAAACGCACCTTTATCCACTGGCGGGAGACGTTCTGAACCTCCCTCCC TTTCGTTCTTTACCCTAAATCACCTTTTTCTTCCTCCTTTCCGCCCGATGCTGTCTCTTGAACTTCTCGTTCTTA CGCTAACGGCACTAACCGCCCCTGTTCTCGCCCATCGCGACTACAACAATGTTCACGCCCGACGGTTTGGTCTAA CCCCTCGGGACTACATCACCCCCGAAACACGACTCGAATCCTACGACTATATCATTGCCGGAGGAGGTCTTGCAG GACTGGTACTTGCTTCCAAGCTCTCGGCTGATGGGGAAACCACTGTTCTCGTCCTCGAGGCTGGAGGAACAGGCG ACGAGCAGAGAGAGACTATTGGTGCGTCCCTTCTGAAATAACATCCTTGAATTATCAACTAACGTCCTCTAGACC GTCCTGGCTTGACATACTTCCGCTCTCTCCTCAACACCGAGCACGATTATGCCTACAAGACCACCCCTCAGGACG GTGCTGGTGGTCGTATCATGAACTGGCCTCGTGGAAAGGTGAGCTTCGATGCATCCTGGTACGTTAATGCTTCTT GCTAACTAAGGCTAGATCCTTGGAGGCTCTTCTGCGGTTAACGGCATGTATCTCGTTCGACCCAACGCGCTCGAA GTTAATGCCATGCACGAGATGAACGCCGATAACGACGCCAAGGAATATGCTGACGCGTGGACCTGGGACTCCCTT CTCGAAGCTATGAAGCAGGGCGAGACCTTCACCCCTCCCACCCAGGAAGCCCTCGATGTTGCGGGCATGCGCTTC AACCCGGACAGCCATGGCTCCAGCGGACCTCTGCAGGCCACTTACTCTGCCTAGTGAGTGCCAGCCATATGTGCA TCTGAAGCTTTGTCTAACCCCTTTCCAGCATGGTCCCCATCTCTAGTGCCTGGCTCCCGACCCTCGAGGCTGCTG GTGTTCCCATTTCCAATGACGCCTACTCCGGAGACAACGTTGGCGGCTTCTTCAGTATCATGGCGATCAACCCCT CCAACTGGACTCGATCATACGCCAAGTCTGCCTATATCGACAACCTTCCTCCCCGCTCCAACCTCCACATCCTCA CCAACAGTGCCGTCACCCGTATCCTCTTTGCCGACAATGCTCAAGATGGCCTCCAGGTCGCGAGCGGAGTCGAGT TCGCCAGCGATGCCAATGATACTCCCAAGACTGTTCTCGCCAACAAGGAAGTGATTCTCGCTGGTGGTGCCGTCG GCAGCCCACACATGTTGTTGGTCAGCGGTGTTGGTGACAGGGCACTGCTTGAGAGCTTGGACATCCCCGTTCGTG TGGACCTCCCTGGTGTTGGCCACCACATGCAAGACCATTTGGTGAGTTCCTTTGTTCATCATTCCTTACTGACAC ATCTGACGGGTACCCTAGGCCGTCAGCGTCGCCTGGAGCACTGAGGAAGACACCCAAGGCACCATCTTCGAATCC GGATCCGAGTTCTCTGTAAGTCGCAACGTTTCCATCGCTGTGCACATCGACTCATCGTTGCCATTAGCAATCCCC CGAGTTCCTTTCCTTCGTAAACTCCGGTACTTCCTATGTCAACGGACGATTCCTGTTCGACGGCCAGGAGCAATT CGACGCCTACCTCCAAGGTCTCCGTGACACCTTCAGCAGCGCCGATGATCTCGCTCCCCTCCTCCCAGGCACCAG CGACGAGGTTGATGAGGGTTACAGGACCATCTACCAGACTTTGGTCGACAAGGTCTACCCCGAGGCTGGCCTGGT CGAGATGCTTTACAGCATCAACGCTGCCGGTCGCATCATCGTCCAGGCTGCAATCCAGACCCCTCTGAGCCAAGG CCGCCTCTCCATCAACTCGACCTCGATCTTCGACCCGCCCGTCGTTGACCCCAAGTACTTCTCCCACCCCGGCGA TATCGTCCTCATGCGCCAAGGTGTGAAGCAGGTCCGCCGCATGTCGACCCTCTCACCCCTCCGTGATATCCTTGG CGACGAGCTCAGCCCCGGTCCTGATGTCCAGTCCGACGAGGACATCGAGAACTGGATCAGGCAGAATGCCAACAC CGAGTTCCACCCCGGCTGCACTTGCGTGATGTTGCCCCGCGACAAGGGCGGTGTTATCGACCAGTCCCTCAAGGT TCACGGTACCGCCAACCTCCGCGTTATTGATGGATCCATCTTCCCTCTCTCTATGTCCGCTCACGTATGTTCCCT CGTTACCATGTGCTCCGTTTACAATGCTGATTGACCTTTGCTCTTCAGTTGATGGCCCCGATCTACGGTGTCGCC GAGAAGGCTGCCGAACTCATCCTCAACCCTCCCTCGTCATCCGGCGGTGGCTCTGGTGGCAAGGGCGGCTCCTCT GGTGGCAAAGGTGGATCCGGCAAGAACGACGAAGATGACGACGAGAGCGCAGCTGCTGGTCTTTCCAGCTCGCTC TTCATCACTGTCGCTGGTTCCCTCTTCACCGCTCTCCTCGGCGCTTTCTAAAGCTCCTCCTGTCTGACTGACTGA CTGACTAACTAACGATATGGACGTTCTATGACCCTTCGGATTCCCATTGCTGGATTTGTTTCTTGACCACGACAC TCATTTGGCTGTCTTATGGACTTGACTTGATACTCGTATATCTTTTCGTTCATTCCTAGCCTAATGAACAGGGAC TTTCCATGCAACCTTTTGATACCATCGCTGTAGATGACGCTGTTATATTAGAGAAAGATGTAATACTGTTGCAAA GTAGTTACCCCCCTGTGTAGCATAGGAATGAACGGACTAGTGTTTGATTTTTGG |
| Length | 2829 |