CopciAB_40535
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_40535 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 40535 |
| Uniprot id | Functional description | Phytanoyl-CoA dioxygenase (PhyH) | |
| Location | scaffold_4:2892324..2894096 | Strand | - |
| Gene length (nt) | 1773 | Transcript length (nt) | 1124 |
| CDS length (nt) | 918 | Protein length (aa) | 305 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Aspergillus nidulans | AN4647 |
| Neurospora crassa | NCU04228 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_116635 | 80.6 | 1.2E-165 | 514 |
| Agrocybe aegerita | Agrae_CAA7260602 | 80.8 | 1.627E-165 | 514 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_196096 | 80.6 | 5.685E-165 | 512 |
| Pleurotus eryngii ATCC 90797 | Pleery1_1460039 | 77.1 | 1.768E-155 | 485 |
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB16916 | 78.4 | 2.262E-154 | 482 |
| Lentinula edodes NBRC 111202 | Lenedo1_1103790 | 76 | 2.529E-151 | 473 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_17934 | 75 | 6.107E-150 | 469 |
| Grifola frondosa | Grifr_OBZ68101 | 75.9 | 6.181E-149 | 466 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1013434 | 76.4 | 4.492E-143 | 449 |
| Pleurotus ostreatus PC9 | PleosPC9_1_50162 | 73.5 | 4.815E-142 | 446 |
| Flammulina velutipes | Flave_chr07AA00306 | 70 | 3.756E-137 | 432 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_127165 | 71.8 | 8.291E-137 | 431 |
| Schizophyllum commune H4-8 | Schco3_2631857 | 60.7 | 6.701E-124 | 394 |
| Auricularia subglabra | Aurde3_1_158341 | 66.1 | 2.538E-121 | 387 |
| Lentinula edodes B17 | Lened_B_1_1_2286 | 71.6 | 7.355E-50 | 179 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_40535.T0 |
| Description | Phytanoyl-CoA dioxygenase (PhyH) |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| Pfam | PF05721 | Phytanoyl-CoA dioxygenase (PhyH) | IPR008775 | 13 | 264 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR008775 | Phytanoyl-CoA dioxygenase-like |
GO
| Go id | Term | Ontology |
|---|---|---|
| No records | ||
KEGG
| KEGG Orthology |
|---|
| No records |
EggNOG
| COG category | Description |
|---|---|
| I | Phytanoyl-CoA dioxygenase (PhyH) |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| AA | AA3 | AA3_2 |
Transcription factor
| Group |
|---|
| No records |
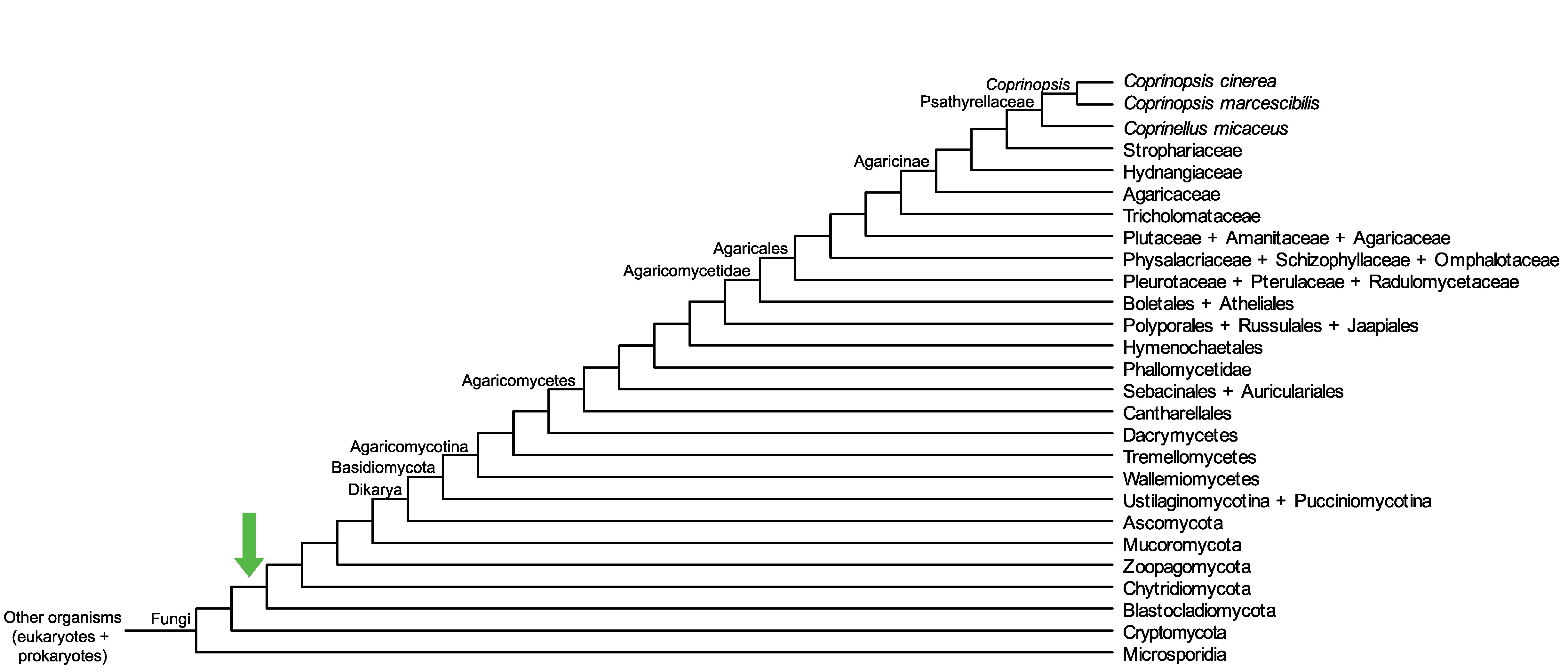
Conservation of CopciAB_40535 across fungi.
Arrow shows the origin of gene family containing CopciAB_40535.
Protein
| Sequence id | CopciAB_40535.T0 |
|---|---|
| Sequence |
>CopciAB_40535.T0 MPEYLSQEQIDQFNKNGYLVLPSFLSEEETSALRKRSKELLDEFDPSTHPLTKFTTGDDDHVGDDYFLTSGDKIR YFLETDAVDGDGKLNREKQKAVNKIGHGLHELDPLFRKVTLENERIRAVVRDLKFHRDPVALQSMVITKQPQIGG EVPEHNDSTFLYTDPPSALGFWIALEKCTESNGALSFLPGSHKTTPITKRFVRLGPGKGTGFETLVSPEEEKRVA EEGKKGEYTLETCNPGDLVLIHGSVLHKSERNTSPNTRFAYTFHMIESPPYANYDEKNWLQPTPEMPFSRILPPG EINVF |
| Length | 305 |
Coding
| Sequence id | CopciAB_40535.T0 |
|---|---|
| Sequence |
>CopciAB_40535.T0 ATGCCAGAATATCTCAGTCAAGAACAGATTGACCAGTTCAATAAGAACGGATATCTCGTTCTGCCGTCATTCTTG TCGGAAGAAGAGACATCCGCGTTGCGGAAGCGCTCTAAGGAGCTGCTGGACGAATTTGACCCTTCTACCCACCCT CTGACCAAGTTCACGACAGGAGACGATGACCACGTCGGAGACGACTATTTCCTGACGTCTGGTGACAAGATACGC TATTTCTTGGAAACGGACGCGGTGGATGGAGATGGAAAGTTGAACAGGGAAAAGCAGAAAGCTGTGAATAAGATT GGACATGGCTTGCACGAGCTTGACCCTCTCTTCCGGAAAGTGACTTTGGAGAATGAACGCATTAGAGCTGTGGTT AGGGATTTGAAGTTCCATCGGGATCCTGTTGCTCTCCAGTCGATGGTAATCACGAAGCAGCCGCAGATTGGTGGC GAAGTGCCTGAGCATAATGACTCGACATTCCTGTACACAGACCCACCATCCGCCCTCGGCTTCTGGATCGCCCTC GAGAAATGCACAGAATCCAACGGAGCCCTGTCTTTCCTTCCCGGATCGCACAAGACCACGCCAATCACCAAACGT TTCGTTCGTCTCGGGCCTGGAAAGGGTACGGGGTTCGAGACGTTGGTCTCACCCGAGGAAGAGAAACGTGTGGCT GAGGAGGGCAAGAAGGGGGAATATACCTTGGAGACTTGCAATCCAGGCGATTTGGTCCTCATCCATGGCTCGGTG CTGCACAAATCGGAGAGGAACACGAGTCCCAATACGAGGTTTGCGTATACCTTCCATATGATCGAGTCGCCGCCC TATGCAAATTATGACGAGAAGAATTGGTTGCAGCCTACACCCGAGATGCCGTTCTCGAGGATCCTGCCTCCGGGG GAGATCAACGTGTTCTAG |
| Length | 918 |
Transcript
| Sequence id | CopciAB_40535.T0 |
|---|---|
| Sequence |
>CopciAB_40535.T0 ACTTTCAACCACCAAGTGCCGTCCTTGTGCATAGTCTCTGCTTTCCATCTCAACAATGCCAGAATATCTCAGTCA AGAACAGATTGACCAGTTCAATAAGAACGGATATCTCGTTCTGCCGTCATTCTTGTCGGAAGAAGAGACATCCGC GTTGCGGAAGCGCTCTAAGGAGCTGCTGGACGAATTTGACCCTTCTACCCACCCTCTGACCAAGTTCACGACAGG AGACGATGACCACGTCGGAGACGACTATTTCCTGACGTCTGGTGACAAGATACGCTATTTCTTGGAAACGGACGC GGTGGATGGAGATGGAAAGTTGAACAGGGAAAAGCAGAAAGCTGTGAATAAGATTGGACATGGCTTGCACGAGCT TGACCCTCTCTTCCGGAAAGTGACTTTGGAGAATGAACGCATTAGAGCTGTGGTTAGGGATTTGAAGTTCCATCG GGATCCTGTTGCTCTCCAGTCGATGGTAATCACGAAGCAGCCGCAGATTGGTGGCGAAGTGCCTGAGCATAATGA CTCGACATTCCTGTACACAGACCCACCATCCGCCCTCGGCTTCTGGATCGCCCTCGAGAAATGCACAGAATCCAA CGGAGCCCTGTCTTTCCTTCCCGGATCGCACAAGACCACGCCAATCACCAAACGTTTCGTTCGTCTCGGGCCTGG AAAGGGTACGGGGTTCGAGACGTTGGTCTCACCCGAGGAAGAGAAACGTGTGGCTGAGGAGGGCAAGAAGGGGGA ATATACCTTGGAGACTTGCAATCCAGGCGATTTGGTCCTCATCCATGGCTCGGTGCTGCACAAATCGGAGAGGAA CACGAGTCCCAATACGAGGTTTGCGTATACCTTCCATATGATCGAGTCGCCGCCCTATGCAAATTATGACGAGAA GAATTGGTTGCAGCCTACACCCGAGATGCCGTTCTCGAGGATCCTGCCTCCGGGGGAGATCAACGTGTTCTAGGT GCTCCTTCGACAACGGTCCTGCGGGTGGTAGGATATGAAGAAGCAAGTGACGGAATCTTTTGTGTTACAACAGTT TGGTGTTAGCGGTATAGCAACAGTGTGGTATCTCTGTGTTAAATAGGTATTATCTAAATGGAATCAGCGTCACG |
| Length | 1124 |
Gene
| Sequence id | CopciAB_40535.T0 |
|---|---|
| Sequence |
>CopciAB_40535.T0 ACTTTCAACCACCAAGTGCCGTCCTTGTGCATAGTCTCTGCTTTCCATCTCAACAATGCCAGAATATCTCAGTCA AGAACAGATTGACCAGGCATGCAACGTATTCCCTGTTGGATCAATGGACGACTGACTCTCGACTCGGATTTGCAG TTCAATAAGAACGGGTATGGTTCGTCTGGTAGTTGATACTGGAGAACGTCTGAACTGATGTAAAGCGAAGATATC TCGTTCTGCCGTCATTCTTGTCGGAAGAAGAGACATCCGCGTTGCGGAAGCGCTCTAAGGAGCTGCTGGACGAAT TTGACCCTTCTACCCACCCTCTGGTATGATCCGCCGACGTACTTCGTCGACATGAACTCACATTTAAGTCTATGG GTTTGCAGACCAAGTTCACGACAGGAGACGATGACCACGTCGGAGACGACTATTTCCTGACGTCTGGTGACAAGA TGTCAGAGTTCATCTACCTCGATTATTCGAAAGTCTCTGCTGACGCGGGTTGTAGACGCTATTTCTTGGAAACGG ACGCGGTGGATGGAGATGGAAAGTTGAACAGGGAAAAGCAGAAAGCTGTGAATAAGATTGGACATGGTGCGTTTA TCCGATCCTTGCTTTGTCCATTGCTGGTTACCTGACCCGTCCCTACGTTGAATCCAAGGCTTGCACGAGCTTGAC CCTCTCTTCCGGAAAGTGACTTTGGAGAATGAACGCATTAGAGCTGTGGTTAGGGATTTGAAGTTCCATCGGGAT CCTGTTGGTATGTCTGCGTCACCAAGGCAAGGACGAATGTAACTGATGCGGATGTCTTGGACGGAATTCAGCTCT CCAGTCGATGGTAATCACGAAGCAGCCGCAGATTGGTGGCGAAGGTACGCTCCTTCTTGTGAGGTTGCTCGAAAT TGCGATGCTGACCTCAAGGGATGGACTGTAGTGCCTGAGCATAATGACTCGTAGGTAACCTTTCTCTCACCCTGC GCAAATCAGTGCATTGAATTCGCATCCTTCGCAGGACATTCCTGGTAAGCTCTCAACATCCCGATCTATCTTCTC CCCAGCCCCTCAGGCTTACTTAACAACACACTCCGACACCTCCAAACAGTACACAGACCCACCATCCGCCCTCGG CTTCTGGATCGCCCTCGAGAAATGCACAGAATCCAACGGAGCCCTGTCTTTCCTTCCCGGATCGCACAAGACCAC GCCAATCACCAAACGTTTCGTTCGTCTCGGGCCTGGAAAGGGTACGGGGTTCGAGACGTTGGTCTCACCCGAGGA AGAGAAACGTGTGGCTGAGGAGGGCAAGAAGGGGGAATATACCTTGGAGACTTGCAATCCAGGTGGGTCTTTCTT ATCTCGGCTGTACGGGTTGAAGTTGTAGCTGACGATGATTGGGTGAATGGACGAATTTTCACATTGTTGTTGTAG GCGATTTGGTCCTCATCCATGGCTCGGTGCTGCACAAATCGGAGAGGAACACGAGTCCCAATACGAGGTTTGCGT ATACCTTCCATATGATCGAGTCGCCGCCCTATGCAAATTATGACGAGAAGAATTGGTTGCAGCCTACACCCGAGA TGCCGTTCTCGAGGATCCTGCCTCCGGGGGAGATCAACGTGTTCTAGGTGCTCCTTCGACAACGGTCCTGCGGGT GGTAGGATATGAAGAAGCAAGTGACGGAATCTTTTGTGTTACAACAGTTTGGTGTTAGCGGTATAGCAACAGTGT GGTATCTCTGTGTTAAATAGGTATTATCTAAATGGAATCAGCGTCACG |
| Length | 1773 |