CopciAB_429295
Coprinopsis cinerea A43mutB43mut pab1-1 #326
General data
| Systematic name | CopciAB_429295 | Strain | Coprinopsis cinerea Okayama 7 |
|---|---|---|---|
| Standard name | - | Synonyms | 429295 |
| Uniprot id | Functional description | P21-Rho-binding domain | |
| Location | scaffold_6:1748394..1752180 | Strand | + |
| Gene length (nt) | 3787 | Transcript length (nt) | 3414 |
| CDS length (nt) | 2340 | Protein length (aa) | 779 |
Reciprocal best hits in model fungi
| Strain name | Gene / Protein name |
|---|---|
| Schizosaccharomyces pombe | shk1 |
| Aspergillus nidulans | AN2067_ste20 |
| Saccharomyces cerevisiae | YHL007C_STE20 |
| Neurospora crassa | stk-4 |
Orthologs in mushroom models
| Strain name | Gene / Protein name | Pident | E-value | Bits |
|---|---|---|---|---|
| Hypsizygus marmoreus strain 51987-8 | Hypma_RDB20838 | 72.7 | 0 | 1121 |
| Pleurotus eryngii ATCC 90797 | Pleery1_135859 | 68.9 | 0 | 1026 |
| Lentinula edodes NBRC 111202 | Lenedo1_1163071 | 68.7 | 0 | 1001 |
| Lentinula edodes W1-26 v1.0 | Lentinedodes1_21726 | 68.7 | 0 | 1001 |
| Agaricus bisporus var bisporus (H97) | Agabi_varbisH97_2_205902 | 76.2 | 0 | 999 |
| Agaricus bisporus var. burnettii JB137-S8 | Agabi_varbur_1_118842 | 76.2 | 0 | 999 |
| Ganoderma sp. 10597 SS1 v1.0 | Gansp1_49456 | 65.4 | 2.47E-304 | 996 |
| Flammulina velutipes | Flave_chr06AA00793 | 66.2 | 4.436E-304 | 978 |
| Pleurotus ostreatus PC9 | PleosPC9_1_106676 | 65.6 | 7.273E-304 | 957 |
| Pleurotus ostreatus PC15 | PleosPC15_2_1057192 | 72.2 | 3.074E-291 | 906 |
| Schizophyllum commune H4-8 | Schco3_2699248 | 64.5 | 2.17E-290 | 901 |
| Lentinula edodes B17 | Lened_B_1_1_14028 | 82.2 | 1.683E-229 | 724 |
| Auricularia subglabra | Aurde3_1_1314578 | 68.3 | 1.231E-200 | 641 |
| Grifola frondosa | Grifr_OBZ73737 | 52.8 | 1.213E-155 | 509 |
Expression
| Name | Summary | Attribution | Assay type | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbon metabolism | Transcriptional changes along alternative morphogenetic paths of C. cinerea | Xie et al. 2020 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex carbon sources 1 (long incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Coprinopsis challenged with bacteria | Transcriptional response to Bacillus subtilis and Escherichia coli. | Kombrink et al. 2018 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 1 | Time series expression profiling of basidiospore and oidium germination | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Early development 2 | Time series expression profiling of mycelial growth, light induction and fruiting body initiation. | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fruiting body development | RNA-Seq analysis of fruiting body development | Krizsan et al. 2019 | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Nitrogen sources (12h incubation) | Expression profiling on diverse nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Plant biomass (12h incubation) | Expression profiling on diverse plant biomasses as carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon and nitrogen sources | Expression profiling on diverse complex carbon and nitrogen sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Simple and complex carbon sources (12h incubation) | Expression profiling on diverse complex carbon sources. | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Starvation induced by water agar transfer | Expression profiling on starvation induced by transfer to water agar | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Straw time series | Time series expression profiling on straw as a carbon source | Hegedus et al. | RNA-Seq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 1 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Stress conditions 2 | Expression profiling under various stress conditions | Hegedus et al. | QuantSeq | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
General data
| Systematic name | - |
|---|---|
| Protein id | CopciAB_429295.T0 |
| Description | P21-Rho-binding domain |
Annotation summary
Conserved domains
| Analysis | Signature accession | Signature description | InterPro Accession | Start | End |
|---|---|---|---|---|---|
| CDD | cd01093 | CRIB_PAK_like | IPR033923 | 235 | 280 |
| CDD | cd06614 | STKc_PAK | - | 504 | 757 |
| Pfam | PF00786 | P21-Rho-binding domain | IPR000095 | 236 | 294 |
| Pfam | PF00069 | Protein kinase domain | IPR000719 | 505 | 756 |
SignalP
| Prediction | Start | End | Score |
|---|---|---|---|
| No records | |||
Transmembrane domains
| Domain n | Start | End | Length |
|---|---|---|---|
| No records | |||
InterPro
| Accession | Description |
|---|---|
| IPR036936 | CRIB domain superfamily |
| IPR000719 | Protein kinase domain |
| IPR000095 | CRIB domain |
| IPR033923 | p21 activated kinase binding domain |
| IPR008271 | Serine/threonine-protein kinase, active site |
| IPR011009 | Protein kinase-like domain superfamily |
| IPR017441 | Protein kinase, ATP binding site |
GO
| Go id | Term | Ontology |
|---|---|---|
| GO:0004672 | protein kinase activity | MF |
| GO:0005524 | ATP binding | MF |
| GO:0006468 | protein phosphorylation | BP |
| GO:0004674 | protein serine/threonine kinase activity | MF |
KEGG
| KEGG Orthology |
|---|
| K04409 |
EggNOG
| COG category | Description |
|---|---|
| T | P21-Rho-binding domain |
CAZy
| Class | Family | Subfamily |
|---|---|---|
| No records | ||
Transcription factor
| Group |
|---|
| No records |
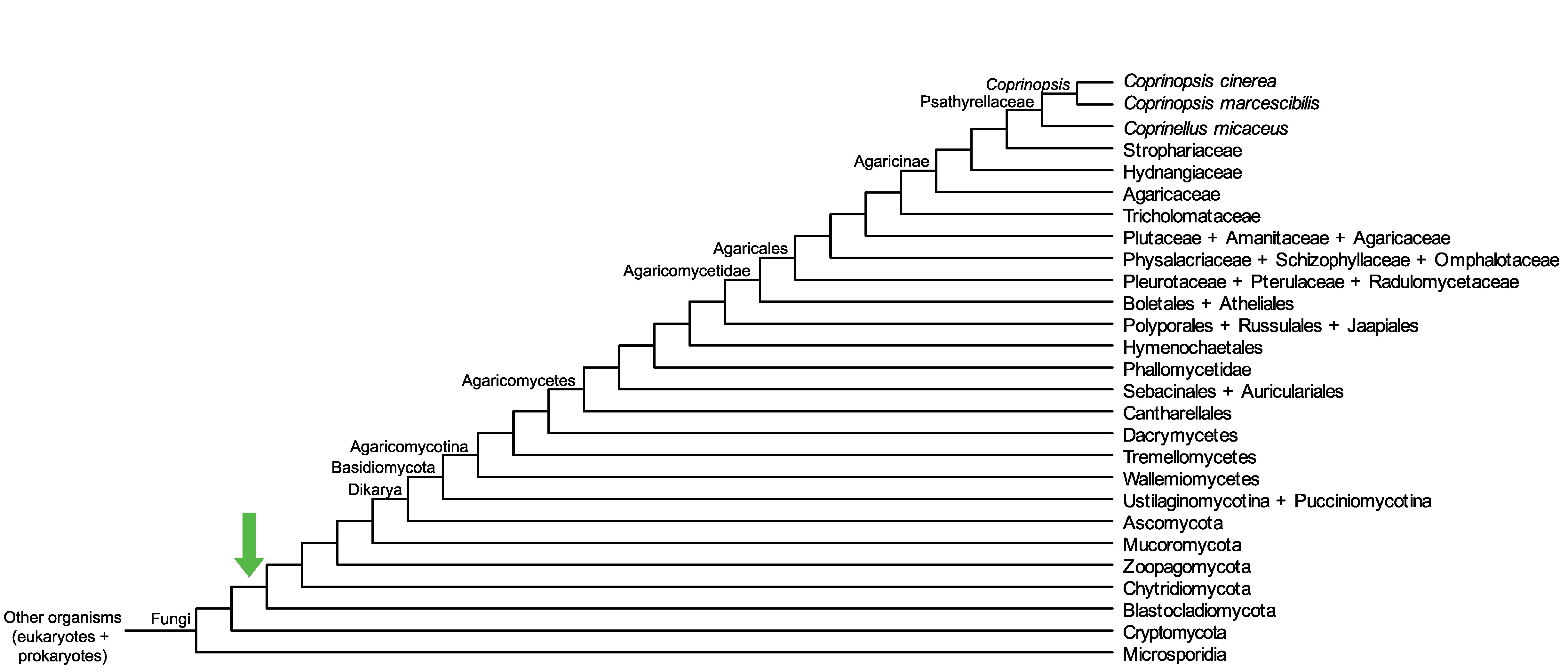
Conservation of CopciAB_429295 across fungi.
Arrow shows the origin of gene family containing CopciAB_429295.
Protein
| Sequence id | CopciAB_429295.T0 |
|---|---|
| Sequence |
>CopciAB_429295.T0 MSSPHTAYNQRPKEPRRQSKITVEYHRPSRESIQEYNALLADANLHAAEAAKYIPSRAPPPPPTNSQRAKTPTDQ QRTRSSLLHKQNPRARNHSLSSQPHHDPFAANVNSQESSFVPPPSPMATHIYNKPPNGTSPPPRPSRANTANLTD IFPTQSQLAARRLSTPSAAPQVHEDGRFYADPNEAMPHNQNMNDYPLPPPPPPSAPLPARVRSGTKSKKGVLGFM SEVFNPTKRPEISTPYDPVHLTHVGFNSSTGEFTGLPKEWQQLLQESGISRHEQEKNPQAVMEIVKFYQEGHGDV WDKLGNVGAPTPEPYALAPENFTNPRSPPPPPKKQPSQASSVHTQHTQSTPTSVGPTSYRPAPTPPTAATPALDR STSQRTPAKPARPADIGRSNTTRDRRSPAPPANNGLAAGSKSYSKPSPGSSQTDLPLKQSAPGPPAHPPRDRADP RAQNHAPSPAASNLAKVAGVATPRRREKKVDKAKDEDIIKRLQQICTDADPTKLYRNLVKIGQGASGGVYTAYQV GTNMSVAIKQMDLDKQPKKDLIINEILVMRSSRHPNIVNYIDSFLYKNDLWVVMEYMEGGSLTDVVTANLMTEGQ IAAVSRETCQGLEHLHRHGVIHRDIKSDNVLLSMVGDIKLTDFGFCAQISEGHAKRTTMVGTPYWMAPEVVTRKE YGPKVDIWSLGIMAIEMIEGEPPYLNQNPLKALYLIATNGTPTIANPENLSPVFTDYLAKTLEVDAEKRPSATEL LQHPFFKLSEPLRTLAPLIKAAREIAKNK |
| Length | 779 |
Coding
| Sequence id | CopciAB_429295.T0 |
|---|---|
| Sequence |
>CopciAB_429295.T0 ATGTCCTCGCCTCACACCGCATATAACCAACGTCCCAAAGAGCCCCGACGCCAATCAAAAATCACCGTCGAGTAC CACAGGCCGTCCAGGGAATCAATCCAAGAATACAACGCTCTCCTCGCAGACGCAAATTTGCACGCGGCAGAAGCT GCAAAGTACATCCCCAGCAGGGCTCCTCCACCGCCCCCGACAAATTCACAGCGGGCCAAGACTCCCACAGACCAA CAACGCACTCGCTCTTCCCTCCTTCACAAGCAAAATCCCCGCGCTAGAAACCATTCCCTGTCCTCACAACCCCAC CACGACCCGTTTGCCGCCAACGTAAACTCTCAAGAGTCGTCCTTTGTCCCTCCGCCGTCTCCAATGGCCACACAC ATATATAACAAGCCACCCAACGGCACATCGCCGCCCCCTCGCCCATCAAGAGCAAACACCGCAAATCTCACCGAC ATCTTCCCGACCCAGTCCCAGCTCGCAGCTCGTCGTTTGTCCACTCCCTCCGCCGCTCCCCAGGTCCACGAGGAT GGCCGCTTTTACGCAGATCCAAACGAGGCAATGCCCCACAATCAGAACATGAACGATTACCCACTACCGCCTCCC CCGCCCCCAAGTGCTCCCCTGCCAGCGAGAGTACGCAGCGGCACCAAATCCAAGAAGGGGGTCCTCGGTTTCATG TCTGAAGTTTTCAATCCAACGAAACGTCCGGAAATCAGCACGCCTTACGATCCCGTCCATCTCACCCACGTCGGG TTCAACTCATCGACAGGAGAATTCACTGGCTTGCCCAAAGAATGGCAGCAATTGTTGCAGGAGAGCGGCATCTCA CGCCACGAGCAGGAGAAGAATCCTCAGGCAGTCATGGAGATCGTCAAGTTCTATCAGGAAGGCCACGGCGATGTG TGGGACAAGCTGGGTAACGTTGGAGCACCAACACCTGAACCCTATGCTCTAGCCCCAGAGAACTTTACCAACCCT CGATCCCCGCCTCCCCCGCCGAAGAAGCAGCCTTCGCAGGCTTCCTCGGTTCATACCCAACATACCCAATCAACC CCGACCTCAGTGGGACCAACATCTTATCGCCCAGCACCAACGCCTCCAACTGCAGCAACCCCTGCGCTTGATCGT TCCACCTCTCAACGGACGCCAGCGAAACCGGCACGTCCGGCTGATATTGGTCGCTCGAATACCACCCGCGATCGT CGATCTCCTGCTCCCCCAGCTAATAACGGTCTCGCGGCTGGATCCAAATCCTATTCGAAGCCATCACCTGGTTCA TCCCAGACAGACCTTCCTCTCAAGCAGTCCGCCCCTGGTCCCCCAGCTCATCCTCCCAGAGATCGCGCGGATCCC CGGGCCCAGAACCATGCTCCCAGCCCTGCCGCTTCCAACCTTGCCAAGGTCGCCGGCGTTGCTACTCCGCGCAGA AGAGAGAAGAAGGTCGACAAGGCGAAGGATGAAGATATCATCAAGCGGCTGCAGCAAATCTGTACTGATGCTGAT CCTACAAAACTCTACAGAAATCTCGTCAAGATTGGTCAAGGTGCTTCTGGAGGTGTATACACTGCCTATCAAGTG GGCACCAATATGTCCGTGGCTATCAAACAGATGGACTTGGACAAGCAGCCCAAGAAGGACCTTATTATCAACGAA ATTCTCGTCATGCGTTCCTCCCGTCATCCCAACATCGTCAACTACATCGACTCATTCCTGTACAAGAATGATCTC TGGGTCGTGATGGAGTACATGGAGGGCGGTTCCCTGACGGACGTCGTTACCGCCAACCTAATGACGGAAGGCCAA ATCGCCGCCGTCTCGCGCGAGACTTGTCAAGGTCTTGAGCACCTCCATCGACACGGTGTCATCCACCGTGATATC AAGAGCGACAACGTCTTGCTGAGCATGGTCGGTGATATTAAATTGACTGATTTCGGTTTCTGTGCACAAATTTCC GAGGGTCATGCCAAGCGAACAACGATGGTTGGTACGCCATATTGGATGGCACCCGAAGTGGTTACGCGCAAAGAG TACGGACCGAAGGTGGACATTTGGAGTTTGGGTATCATGGCTATTGAGATGATCGAAGGTGAACCGCCATATCTC AACCAGAACCCGCTCAAGGCCCTCTACTTGATTGCCACCAACGGCACGCCGACAATTGCCAACCCTGAAAATCTA TCGCCCGTCTTCACGGACTACTTGGCCAAGACCCTCGAGGTTGATGCTGAGAAACGGCCCTCGGCGACGGAACTG CTTCAGCATCCGTTCTTTAAACTGTCGGAACCTCTTCGAACGCTTGCTCCTCTAATCAAGGCTGCAAGGGAAATC GCGAAGAACAAGTAA |
| Length | 2340 |
Transcript
| Sequence id | CopciAB_429295.T0 |
|---|---|
| Sequence |
>CopciAB_429295.T0 GAACTACTCTTGCACCACCACCACCATCTTCCCATCCCTCTTTCAACATCGTCGTCGTCGTCGTCGTCGTCGTCG TCGTCGACAGCCAACACAGCGATACATAAATAAGCGTGGGCGGCACACGGACCCCTGAACCGTGACCGTGAGGAT ATCGTTGCCTTGGACTTTTGGTCGTCATCATTGATATCCGTTGACAACCAAGATCTTGCAGAAAAGAGGAGAGAT TGAAGGGAGCAGAGGGAGAGTTGCATCGTCGACCATCAGTCTTGAATTTGAAGGCGCTAGCATCGGACGCGTCTC GGTTTTCGACCCTGAAAGGGCCTAATTCCATCATTCGCCGCAATATCGGGTATGTCCTCGCCTCACACCGCATAT AACCAACGTCCCAAAGAGCCCCGACGCCAATCAAAAATCACCGTCGAGTACCACAGGCCGTCCAGGGAATCAATC CAAGAATACAACGCTCTCCTCGCAGACGCAAATTTGCACGCGGCAGAAGCTGCAAAGTACATCCCCAGCAGGGCT CCTCCACCGCCCCCGACAAATTCACAGCGGGCCAAGACTCCCACAGACCAACAACGCACTCGCTCTTCCCTCCTT CACAAGCAAAATCCCCGCGCTAGAAACCATTCCCTGTCCTCACAACCCCACCACGACCCGTTTGCCGCCAACGTA AACTCTCAAGAGTCGTCCTTTGTCCCTCCGCCGTCTCCAATGGCCACACACATATATAACAAGCCACCCAACGGC ACATCGCCGCCCCCTCGCCCATCAAGAGCAAACACCGCAAATCTCACCGACATCTTCCCGACCCAGTCCCAGCTC GCAGCTCGTCGTTTGTCCACTCCCTCCGCCGCTCCCCAGGTCCACGAGGATGGCCGCTTTTACGCAGATCCAAAC GAGGCAATGCCCCACAATCAGAACATGAACGATTACCCACTACCGCCTCCCCCGCCCCCAAGTGCTCCCCTGCCA GCGAGAGTACGCAGCGGCACCAAATCCAAGAAGGGGGTCCTCGGTTTCATGTCTGAAGTTTTCAATCCAACGAAA CGTCCGGAAATCAGCACGCCTTACGATCCCGTCCATCTCACCCACGTCGGGTTCAACTCATCGACAGGAGAATTC ACTGGCTTGCCCAAAGAATGGCAGCAATTGTTGCAGGAGAGCGGCATCTCACGCCACGAGCAGGAGAAGAATCCT CAGGCAGTCATGGAGATCGTCAAGTTCTATCAGGAAGGCCACGGCGATGTGTGGGACAAGCTGGGTAACGTTGGA GCACCAACACCTGAACCCTATGCTCTAGCCCCAGAGAACTTTACCAACCCTCGATCCCCGCCTCCCCCGCCGAAG AAGCAGCCTTCGCAGGCTTCCTCGGTTCATACCCAACATACCCAATCAACCCCGACCTCAGTGGGACCAACATCT TATCGCCCAGCACCAACGCCTCCAACTGCAGCAACCCCTGCGCTTGATCGTTCCACCTCTCAACGGACGCCAGCG AAACCGGCACGTCCGGCTGATATTGGTCGCTCGAATACCACCCGCGATCGTCGATCTCCTGCTCCCCCAGCTAAT AACGGTCTCGCGGCTGGATCCAAATCCTATTCGAAGCCATCACCTGGTTCATCCCAGACAGACCTTCCTCTCAAG CAGTCCGCCCCTGGTCCCCCAGCTCATCCTCCCAGAGATCGCGCGGATCCCCGGGCCCAGAACCATGCTCCCAGC CCTGCCGCTTCCAACCTTGCCAAGGTCGCCGGCGTTGCTACTCCGCGCAGAAGAGAGAAGAAGGTCGACAAGGCG AAGGATGAAGATATCATCAAGCGGCTGCAGCAAATCTGTACTGATGCTGATCCTACAAAACTCTACAGAAATCTC GTCAAGATTGGTCAAGGTGCTTCTGGAGGTGTATACACTGCCTATCAAGTGGGCACCAATATGTCCGTGGCTATC AAACAGATGGACTTGGACAAGCAGCCCAAGAAGGACCTTATTATCAACGAAATTCTCGTCATGCGTTCCTCCCGT CATCCCAACATCGTCAACTACATCGACTCATTCCTGTACAAGAATGATCTCTGGGTCGTGATGGAGTACATGGAG GGCGGTTCCCTGACGGACGTCGTTACCGCCAACCTAATGACGGAAGGCCAAATCGCCGCCGTCTCGCGCGAGACT TGTCAAGGTCTTGAGCACCTCCATCGACACGGTGTCATCCACCGTGATATCAAGAGCGACAACGTCTTGCTGAGC ATGGTCGGTGATATTAAATTGACTGATTTCGGTTTCTGTGCACAAATTTCCGAGGGTCATGCCAAGCGAACAACG ATGGTTGGTACGCCATATTGGATGGCACCCGAAGTGGTTACGCGCAAAGAGTACGGACCGAAGGTGGACATTTGG AGTTTGGGTATCATGGCTATTGAGATGATCGAAGGTGAACCGCCATATCTCAACCAGAACCCGCTCAAGGCCCTC TACTTGATTGCCACCAACGGCACGCCGACAATTGCCAACCCTGAAAATCTATCGCCCGTCTTCACGGACTACTTG GCCAAGACCCTCGAGGTTGATGCTGAGAAACGGCCCTCGGCGACGGAACTGCTTCAGCATCCGTTCTTTAAACTG TCGGAACCTCTTCGAACGCTTGCTCCTCTAATCAAGGCTGCAAGGGAAATCGCGAAGAACAAGTAAAGGACAATT TTCTCTGTGGACAGCACCTTCCATTTTTCTCTCTTTCTTCTTCTCTTTTTTCTCTTCTCTCATTAAATCTGTGTT TGTTCGCGATCGATGATATATCCCCCTCCTCTCCAATTCCCGGTCCGAGTCTCATAGTCCGCATCTAATACCTGG TTGTGAAAACCATTTCTTTTGGTATCTTCTTATGCATTGATGGTTCTCGGTCAACTATATCTCCCTTTCTTCACC CTTCCTATTACCTCCCACTTCAATCTCCTTCGTCGACCAAGATCGTCTGTTTCACCTTACATATATGCCCTTCGT CCTTTCTTTCGGTCGACGCGGTAGTATTTAACTGGCCATAAGAGTCTTAGAATCCGGCACTGGAGGCTTCTTTAA CCCCCCTTTCTCATATTTATTGCGCGCACCCAGGACATCTCGTTTGTTAGATTGTCCTAGGACGGTGTATTCACC TTCACCCTACACCACACACATCTGTACATACCTTTTTTGTTCAACAACCCTCCAGGTCTGGTTTCCTCACGGTCG GTTTTGCAAGACTTTTCTCTCTCGTGTGTGTTCTGCCCCTCTCTGTTTCTGTTATACATTCAATCATCGTAATGA TTTCTCGGTTGGAAGTTTGAAAAAGAAAAAAGACAATGGCTACTCTATGTATATAATACAAAATGCTTGTCCGTG TTTAGTAGATGATGCAATTGATTATTCGACAGACGATGA |
| Length | 3414 |
Gene
| Sequence id | CopciAB_429295.T0 |
|---|---|
| Sequence |
>CopciAB_429295.T0 GAACTACTCTTGCACCACCACCACCATCTTCCCATCCCTCTTTCAACATCGTCGTCGTCGTCGTCGTCGTCGTCG TCGTCGACAGCCAACACAGCGATACATAAATAAGCGTGGGCGGCACACGGACCCCTGAACCGTGACCGTGAGGAT ATCGTTGCCTTGGACTTTTGGTCGTCATCATTGATATCCGTTGACAACCAAGATCTTGCAGAAAAGAGGAGAGAT TGAAGGGAGCAGAGGGAGAGTTGCATCGTCGACCATCAGTCTTGAATTTGAAGGCGCTAGCATCGGACGCGTCTC GGTTTTCGACCCTGAAAGGGCCTAATTCCATCATTCGCCGCAATATCGGGTATGTCCTCGCCTCACACCGCATAT AACCAACGTCCCAAAGAGCCCCGACGCCAATCAAAAATCACCGTCGAGTACCACAGGCCGTCCAGGGAATCAATC CAAGAATACAACGCTCTCCTCGCAGACGCAAATTTGCACGCGGCAGAAGCTGCAAAGTACATCCCCAGCAGGGCT CCTCCACCGCCCCCGACAAATTCACAGCGGGCCAAGACTCCCACAGACCAACAACGCACTCGCTCTTCCCTCCTT CACAAGCAAAATCCCCGCGCTAGAAACCATTCCCTGTCCTCACAACCCCACCACGACCCGTTTGCCGCCAACGTA AACTCTCAAGAGTCGTCCTTTGTCCCTCCGCCGTCTCCAATGGCCACACACATATATAACAAGCCACCCAACGGC ACATCGCCGCCCCCTCGCCCATCAAGAGCAAACACCGCAAATCTCACCGACATCTTCCCGACCCAGTCCCAGCTC GCAGCTCGTCGTTTGTCCACTCCCTCCGCCGCTCCCCAGGTCCACGAGGATGGCCGCTTTTACGCAGATCCAAAC GAGGCAATGCCCCACAATCAGAACATGAACGATTACCCACTACCGCCTCCCCCGCCCCCAAGTGCTCCCCTGCCA GCGAGAGTACGCAGCGGCACCAAATCCAAGAAGGGGGTCCTCGGTTTCATGTCTGAAGTTTTCAATCCAACGAAA CGTCCGGAAATCAGCACGCCTTACGATCCCGTCCATCTCACCCACGTCGGGTTCAACTCATCGACAGGAGAATTC ACTGGCTTGCCCAAAGAATGGCAGCAATTGTTGCAGGAGAGCGGCATCTCACGCCACGAGCAGGAGAAGAATCCT CAGGCAGTCATGGAGATCGTCAAGTTCTATCAGGAAGGCCACGGCGATGTGTGGGACAAGCTGGGTAACGTTGGA GCACCAACACCTGAACCCTATGCTCTAGCCCCAGAGAACTTTACCAACCCTGTCAGTCCCATTTCCGTAACTATA TGCCACATGATAATTAACATCTATTGTTTCGCAGCGATCCCCGCCTCCCCCGCCGAAGAAGCAGCCTTCGCAGGC TTCCTCGGTTCATACCCAACATACCCAATCAACCCCGACCTCAGTGGGACCAACATCTTATCGCCCAGCACCAAC GCCTCCAACTGCAGCAACCCCTGCGCTTGATCGTTCCACCTCTCAACGGACGCCAGCGAAACCGGCACGTCCGGC TGATATTGGTCGCTCGAATACCACCCGCGATCGTCGATCTCCTGCTCCCCCAGCTAATAACGGTCTCGCGGCTGG ATCCAAATCCTATTCGAAGCCATCACCTGGTTCATCCCAGACAGACCTTCCTCTCAAGCAGTCCGCCCCTGGTCC CCCAGCTCATCCTCCCAGAGATCGCGCGGATCCCCGGGCCCAGAACCATGCTCCCAGCCCTGCCGCTTCCAACCT TGCCAAGGTCGCCGGCGTTGCTACTCCGCGCAGAAGAGAGAAGAAGGTCGACAAGGCGAAGGATGAAGATATCAT CAAGCGGCTGCAGCAAATCTGTACTGATGCTGATCCTACAAAACTCTACAGAAATCTCGTCAAGATTGGTCAAGG GTGAGGATCATCGATATGCTATGCTTAATCCAAGTCTTCTGAACTCGCAAACAATAGTGCTTCTGGAGGTGTATA CACTGCCTATCAAGTGGGCACCAATATGTCCGTGGCTATCAAACAGATGGACTTGGACAAGCAGCCCAAGAAGGA CCTTATTATCAACGAAATTCTCGTCATGCGTTCCTCCCGTCATCCCAACATCGTCAACTACATCGACTCATTCCT GTACAAGAATGATCTCTGGGTCGTGATGGAGTACATGGAGGGCGGTTCCCTGACGGACGTCGTTACCGCCAACCT AATGACGGAAGGCCAAATCGCCGCCGTCTCGCGCGAGACTTGTCAAGGTCTTGAGCACCTCCATCGACACGGTGT CATCCACCGTGATATCAAGAGCGACAACGTCTTGCTGAGCATGGTCGGTGATATTAAATTGAGTAAGTGTGTCTA CACCCGTGATGATCCTTGTTTTCTGACCATTGGATATTTTTTGCTTCGCCTCGTCTTCTTCGGCATTTGCCTGGA CCACCCTTTCTAGCTGATTTCGGTTTCTGTGCACAAATTTCCGAGGGTCATGCCAAGCGAACAACGATGGTTGGT ACGCCATATTGGATGGCACCCGAAGTGGTTACGCGCAAAGAGTACGGACCGAAGGTGGACATTTGGAGTTTGGGT ATCATGGCTATTGGTGAGTGTTGTCAATCTAATTTTCCGATCTCCCCGTGTTTTGGCTGACCAGGGTTTGTTGGG TGAACAGAGATGATCGAAGGTGAACCGCCATATCTCAACCAGAACCCGCTCAAGGCCCTCTACTTGATTGCCACC AACGGCACGCCGACAATTGCCAACCCTGAAAATCTATCGCCCGTCTTCACGGACTACTTGGCCAAGACCCTCGAG GTTGATGCTGAGAAACGGCCCTCGGCGACGGAACTGCTTCAGGTGCGCATTTTTTTCATTAACTTTGGTCCACAT GGCCAAGGATTGCACTGCCACGACGTTGGATACTGATGATTGATTGGATTTTTAGCATCCGTTCTTTAAACTGTC GGAACCTCTTCGAACGCTTGCTCCTCTAATCAAGGCTGCAAGGGAAATCGCGAAGAACAAGTAAAGGACAATTTT CTCTGTGGACAGCACCTTCCATTTTTCTCTCTTTCTTCTTCTCTTTTTTCTCTTCTCTCATTAAATCTGTGTTTG TTCGCGATCGATGATATATCCCCCTCCTCTCCAATTCCCGGTCCGAGTCTCATAGTCCGCATCTAATACCTGGTT GTGAAAACCATTTCTTTTGGTATCTTCTTATGCATTGATGGTTCTCGGTCAACTATATCTCCCTTTCTTCACCCT TCCTATTACCTCCCACTTCAATCTCCTTCGTCGACCAAGATCGTCTGTTTCACCTTACATATATGCCCTTCGTCC TTTCTTTCGGTCGACGCGGTAGTATTTAACTGGCCATAAGAGTCTTAGAATCCGGCACTGGAGGCTTCTTTAACC CCCCTTTCTCATATTTATTGCGCGCACCCAGGACATCTCGTTTGTTAGATTGTCCTAGGACGGTGTATTCACCTT CACCCTACACCACACACATCTGTACATACCTTTTTTGTTCAACAACCCTCCAGGTCTGGTTTCCTCACGGTCGGT TTTGCAAGACTTTTCTCTCTCGTGTGTGTTCTGCCCCTCTCTGTTTCTGTTATACATTCAATCATCGTAATGATT TCTCGGTTGGAAGTTTGAAAAAGAAAAAAGACAATGGCTACTCTATGTATATAATACAAAATGCTTGTCCGTGTT TAGTAGATGATGCAATTGATTATTCGACAGACGATGA |
| Length | 3787 |